bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0960_orf1
Length=223
Score E
Sequences producing significant alignments: (Bits) Value
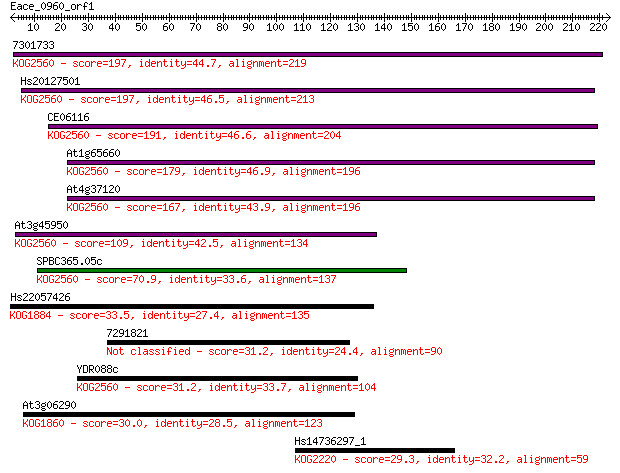
7301733 197 2e-50
Hs20127501 197 2e-50
CE06116 191 8e-49
At1g65660 179 5e-45
At4g37120 167 2e-41
At3g45950 109 4e-24
SPBC365.05c 70.9 2e-12
Hs22057426 33.5 0.37
7291821 31.2 1.5
YDR088c 31.2 1.6
At3g06290 30.0 4.3
Hs14736297_1 29.3 7.0
> 7301733
Length=527
Score = 197 bits (501), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 98/220 (44%), Positives = 144/220 (65%), Gaps = 4/220 (1%)
Query 2 KEFDKTSAPVECTDERSRINTRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLS 61
K D+ P D + RI R+LRIREDTAKYL NLD NSA+YDPK+RSMR++P +
Sbjct 215 KYVDEVDMPGTKVDSKQRITVRNLRIREDTAKYLRNLDPNSAYYDPKTRSMRDNPNPAVP 274
Query 62 QQIKPTFRGDNALLQAGEVKATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKA 121
++ + F G+N + +G+ A QLFAWEA+ G +VH A+PT+LE L KE+++KK
Sbjct 275 EE-EAEFAGENFVRFSGDTTAQATAQLFAWEAHGKGVDVHLLAEPTKLELLQKEYEQKKE 333
Query 122 QIETEKQKNIFDTYGGKEHL-VADTRVLYAQTEAYVEYSKDGRILRGRQRTLTKSKYEED 180
Q ++ + +I + YGG+EHL V +L AQTE Y+EYS+ G++++G ++ +S YEED
Sbjct 334 QFKSSTKTHIVEKYGGEEHLQVPPKSLLLAQTEEYIEYSRSGKVIKGVEKPKARSIYEED 393
Query 181 VLQGSHTSVWGSYYDVATSKWGYACCRLTSFAAECTGRLE 220
V +HT+VWGS+++ +WGY CC+ + C G E
Sbjct 394 VYINNHTTVWGSFWNAG--RWGYKCCKSFIKNSYCVGMQE 431
> Hs20127501
Length=586
Score = 197 bits (500), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 99/215 (46%), Positives = 138/215 (64%), Gaps = 4/215 (1%)
Query 5 DKTSAPVECTDERSRINTRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLSQQI 64
D P + D + RI R+LRIRED AKYL NLD NSA+YDPK+R+MRE+P+ + +
Sbjct 245 DDIDMPGQNFDSKRRITVRNLRIREDIAKYLRNLDPNSAYYDPKTRAMRENPYANAGKNP 304
Query 65 -KPTFRGDNALLQAGEVKATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKAQI 123
+ ++ GDN + G+ + Q QLFAWEAY G VH A PT+LE LYK + KK
Sbjct 305 DEVSYAGDNFVRYTGDTISMAQTQLFAWEAYDKGSEVHLQADPTKLELLYKSFKVKKEDF 364
Query 124 ETEKQKNIFDTYGGKEHLVA-DTRVLYAQTEAYVEYSKDGRILRGRQRTLTKSKYEEDVL 182
+ +++++I + YGG+EHL A +L AQTE YVEYS+ G +++G++R + SKYEEDV
Sbjct 365 KEQQKESILEKYGGQEHLDAPPAELLLAQTEDYVEYSRHGTVIKGQERAVACSKYEEDVK 424
Query 183 QGSHTSVWGSYYDVATSKWGYACCRLTSFAAECTG 217
+HT +WGSY+ +WGY CC + CTG
Sbjct 425 IHNHTHIWGSYW--KEGRWGYKCCHSFFKYSYCTG 457
> CE06116
Length=647
Score = 191 bits (486), Expect = 8e-49, Method: Compositional matrix adjust.
Identities = 95/208 (45%), Positives = 144/208 (69%), Gaps = 7/208 (3%)
Query 15 DERSRINTRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLS-QQIKPT-FRGDN 72
D R+RI R+LRIREDTAKYL NL NS +YDPKSRSMRE+PF ++ ++++ F GDN
Sbjct 230 DSRTRITVRNLRIREDTAKYLYNLAENSPYYDPKSRSMRENPFAGVAGKELEAARFSGDN 289
Query 73 ALLQAGEVKATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKAQIETEKQKNIF 132
+ +GEV A + Q+FAW+A + G H A+PT+LE L KE++K+K+ ++ E QK +
Sbjct 290 FVRYSGEVTAANEAQVFAWQATRGGVYAHSIAEPTKLEALKKEYEKEKSTLKNETQKELL 349
Query 133 DTYGGKEHL--VADTRVLYAQTEAYVEYSKDGRILRGRQRTLTKSKYEEDVLQGSHTSVW 190
D YGG EH+ AD +L AQTE+Y+EY++ G++++G+++ S+++ED+ +HTSV+
Sbjct 350 DKYGGGEHMERPAD-ELLLAQTESYIEYNRKGKVIKGKEKVAISSRFKEDIYPQNHTSVF 408
Query 191 GSYYDVATSKWGYACCRLTSFAAECTGR 218
GS++ +WGY CC + CTG+
Sbjct 409 GSFW--REGRWGYKCCHQFVKNSYCTGK 434
> At1g65660
Length=535
Score = 179 bits (453), Expect = 5e-45, Method: Compositional matrix adjust.
Identities = 92/198 (46%), Positives = 124/198 (62%), Gaps = 6/198 (3%)
Query 22 TRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLSQQIKPTFRGDNALLQAGEVK 81
R+LRIREDTAKYLLNLD+NSA YDPK+RSMREDP K + GDN +G+
Sbjct 235 VRNLRIREDTAKYLLNLDVNSAHYDPKTRSMREDPLPDADPNDK-FYLGDNQYRNSGQAL 293
Query 82 ATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKAQIETEKQKNIFDTYG--GKE 139
+QL + +WEA+ GQ++H A P+Q E LYK Q K +++++ + I D YG E
Sbjct 294 EFKQLNIHSWEAFDKGQDMHMQAAPSQAELLYKSFQVAKEKLKSQTKDTIMDKYGNAATE 353
Query 140 HLVADTRVLYAQTEAYVEYSKDGRILRGRQRTLTKSKYEEDVLQGSHTSVWGSYYDVATS 199
+ +L Q+E VEY + GRI++G++ L KSKYEEDV +HTSVWGSY+
Sbjct 354 DEIP-MELLLGQSERQVEYDRAGRIIKGQEVILPKSKYEEDVHANNHTSVWGSYW--KDH 410
Query 200 KWGYACCRLTSFAAECTG 217
+WGY CC+ + CTG
Sbjct 411 QWGYKCCQQIIRNSYCTG 428
> At4g37120
Length=538
Score = 167 bits (422), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 86/198 (43%), Positives = 123/198 (62%), Gaps = 6/198 (3%)
Query 22 TRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLSQQIKPTFRGDNALLQAGEVK 81
R+LRIREDTAKYLLNLD+NSA YDPK+RSMREDP K + GDN +G+
Sbjct 237 VRNLRIREDTAKYLLNLDVNSAHYDPKTRSMREDPLPDADPNEK-FYLGDNQYRNSGQAL 295
Query 82 ATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKAQIETEKQKNIFDTYG--GKE 139
+Q+ + + EA+ G ++H A P+Q E LYK + K +++T+ + I + YG E
Sbjct 296 EFKQINIHSCEAFDKGHDMHMQAAPSQAELLYKNFKVAKEKLKTQTKDTIMEKYGNAATE 355
Query 140 HLVADTRVLYAQTEAYVEYSKDGRILRGRQRTLTKSKYEEDVLQGSHTSVWGSYYDVATS 199
+ +L Q+E +EY + GRI++G++ + KSKYEEDV +HTSVWGS++
Sbjct 356 GEIP-MELLLGQSERQIEYDRAGRIMKGQEVIIPKSKYEEDVHANNHTSVWGSWW--KDH 412
Query 200 KWGYACCRLTSFAAECTG 217
+WGY CC+ T + CTG
Sbjct 413 QWGYKCCQQTIRNSYCTG 430
> At3g45950
Length=385
Score = 109 bits (273), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 57/134 (42%), Positives = 80/134 (59%), Gaps = 1/134 (0%)
Query 3 EFDKTSAPVECTDERSRINTRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLSQ 62
+F K V TD S+ R+LRIRED AKYLLNLD+NSA+YDPKSRSMREDP +
Sbjct 206 DFAKVKKRVRTTDGGSKGTVRNLRIREDPAKYLLNLDVNSAYYDPKSRSMREDPLPYTDP 265
Query 63 QIKPTFRGDNALLQAGEVKATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKAQ 122
K R DN +G+ +Q +++ EA+ GQ++H A P+Q E YK + K +
Sbjct 266 NEKFCLR-DNQYRNSGQAIEFKQQNMYSCEAFDKGQDIHMQAAPSQAELCYKRVKIAKEK 324
Query 123 IETEKQKNIFDTYG 136
+ ++++ I YG
Sbjct 325 LNSQRKDAIIAKYG 338
> SPBC365.05c
Length=379
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 46/144 (31%), Positives = 73/144 (50%), Gaps = 17/144 (11%)
Query 11 VECTDERSRINTRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLSQQIKPTFRG 70
V +++ + I T LR+RED YL D + Y+PKSRSMR++ H+
Sbjct 207 VSGSEDSASITTPSLRMREDVVAYL-RADNKNLQYEPKSRSMRDETGYHMV--------- 256
Query 71 DNALLQAGEVKAT-------QQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKAQI 123
D++ AG VKA+ ++LQ+FAWEA + G VH A PT E +++++ +
Sbjct 257 DDSSGGAGFVKASGGEKEDFEKLQMFAWEAERSGTRVHVVANPTAGELEFRKNKASRMTT 316
Query 124 ETEKQKNIFDTYGGKEHLVADTRV 147
+ ++I D YG V D +
Sbjct 317 QKHIDQSILDRYGDGTSKVKDKKA 340
> Hs22057426
Length=2407
Score = 33.5 bits (75), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 37/142 (26%), Positives = 58/142 (40%), Gaps = 22/142 (15%)
Query 1 LKEFDKTSAPVECTDERSRINTRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHL 60
L + + S P+ + + I R L DT + L N ++P S+S D F HL
Sbjct 520 LNGWGRCSIPIHALE--AGIENRQL----DTVNFFLKSKEN--LFNPSSKSSVSDQFDHL 571
Query 61 SQQIKPTFRGDNALLQAGEVKATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKK 120
S + R L+ A ++ + E+Y Q+ HF+ Q L + +Q K+
Sbjct 572 SSHL--YLRNVEELIPALDLLCSA-----IRESYSEPQSKHFSEQLLNLTLSFLNNQIKE 624
Query 121 AQIETE-------KQKNIFDTY 135
I TE K NI +Y
Sbjct 625 LFIHTEELDEHLQKGVNILTSY 646
> 7291821
Length=280
Score = 31.2 bits (69), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 22/93 (23%), Positives = 43/93 (46%), Gaps = 5/93 (5%)
Query 37 NLDLNSAFYDPKSRSMREDPFKHLSQQIKPTFRGDNALLQAGEVKATQQLQLFAWEAYKH 96
N+ L A DP+ + +D ++ + PTF ++ Q +++ T+ L +
Sbjct 4 NILLQLAAEDPQRDAYPKDTVCAIASALHPTFPSNSEKFQKDQLQPTESLLPKSASPNPR 63
Query 97 GQNVHFNAQPTQLEFLYK---EHQKKKAQIETE 126
GQ++ +N P +E ++K EH Q+ E
Sbjct 64 GQSITYN--PPSMEKIWKKVCEHHDVPEQVANE 94
> YDR088c
Length=382
Score = 31.2 bits (69), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 35/112 (31%), Positives = 55/112 (49%), Gaps = 14/112 (12%)
Query 26 RIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLSQQIKPTFR----GD----NALLQA 77
R+RED A YL +++ + YDPKSR + + + ++ K FR G+ N L Q
Sbjct 247 RLREDKAAYLNDINSTESNYDPKSRLYKTETLGAVDEKSK-MFRRHLTGEGLKLNELNQF 305
Query 78 GEVKATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKAQIETEKQK 129
A + E + Q+V A PT+ E+L +KK+ Q ET++ K
Sbjct 306 ARSHAKEMGIRDEIEDKEKVQHV-LVANPTKYEYL----KKKREQEETKQPK 352
> At3g06290
Length=1713
Score = 30.0 bits (66), Expect = 4.3, Method: Composition-based stats.
Identities = 35/138 (25%), Positives = 61/138 (44%), Gaps = 20/138 (14%)
Query 6 KTSAPVECTDERSRINTRDLRIREDTAKYLLNL---DLNSAFYD------PKSRSMRED- 55
K S+P T ER I R + I ++T +YLL+L N F + R++R D
Sbjct 524 KVSSP---TAEREAILIRPMPILQNTMEYLLSLLDRPYNENFLGMYNFLWDRMRAIRMDL 580
Query 56 PFKHLSQQIKPTFRGDNALLQAGEVKATQQLQLFAWEAYKHGQNVHFNAQ---PTQLEF- 111
+H+ Q T +++ + + + E + G + H N + T +E
Sbjct 581 RMQHIFNQEAITLL--EQMIRLHIIAMHELCEYTKGEGFSEGFDAHLNIEQMNKTSVELF 638
Query 112 -LYKEHQKKKAQIETEKQ 128
+Y +H+KK + TEK+
Sbjct 639 QMYDDHRKKGITVPTEKE 656
> Hs14736297_1
Length=1189
Score = 29.3 bits (64), Expect = 7.0, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 32/59 (54%), Gaps = 10/59 (16%)
Query 107 TQLEFLYKEHQKKKAQIETEKQKNIFDTYGGKEHLVADTRVLYAQTEAYVEYSKDGRIL 165
++++ L++E KK Q++ ++N L A RVL A TEA V+Y+ R+L
Sbjct 596 SEMKKLFEEQLKKYDQLKVYLEQN----------LAAQDRVLCALTEANVQYAAVRRVL 644
Lambda K H
0.316 0.131 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4255059914
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40