bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0954_orf1
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
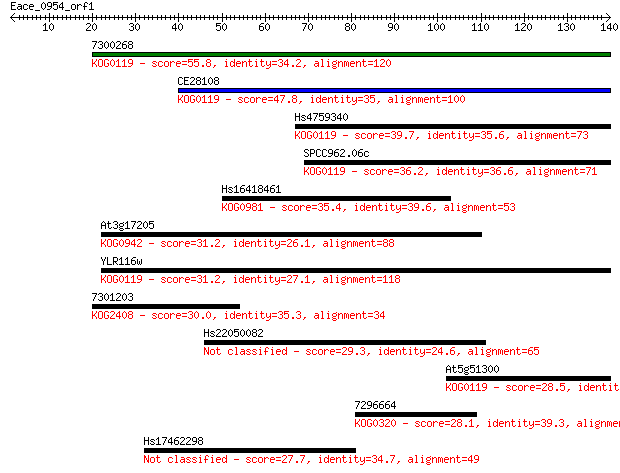
7300268 55.8 3e-08
CE28108 47.8 7e-06
Hs4759340 39.7 0.002
SPCC962.06c 36.2 0.021
Hs16418461 35.4 0.031
At3g17205 31.2 0.61
YLR116w 31.2 0.61
7301203 30.0 1.4
Hs22050082 29.3 2.7
At5g51300 28.5 4.0
7296664 28.1 5.5
Hs17462298 27.7 6.3
> 7300268
Length=663
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 41/121 (33%), Positives = 58/121 (47%), Gaps = 13/121 (10%)
Query 20 FISFLFNSFNKMSLEQLMALVPLEAAMTHKEKGRKSRWEKSKKSADGSRWGSETDKPFLP 79
F SF FNS N A + +++ RKSRW GSE DK F+P
Sbjct 242 FASF-FNSQNSNDSTSNGAFDNSADSAAERKRKRKSRW-----------GGSENDKTFIP 289
Query 80 PPFIDLPIGMTPLQVDRFLREQRLEELYRKLNNGILEFF-DADIRPPSPPPVYDKNGGRV 138
LP + P Q + +L + ++EE+ RKL G L + + R PSP P+Y +G R+
Sbjct 290 GMPTILPSTLDPAQQEAYLVQFQIEEISRKLRTGDLGITQNPEERSPSPEPIYSSDGKRL 349
Query 139 N 139
N
Sbjct 350 N 350
> CE28108
Length=699
Score = 47.8 bits (112), Expect = 7e-06, Method: Composition-based stats.
Identities = 35/107 (32%), Positives = 48/107 (44%), Gaps = 21/107 (19%)
Query 40 VPLEA-------AMTHKEKGRKSRWEKSKKSADGSRWGSETDKPFLPPPFIDLPIGMTPL 92
VP+EA A+ +K R+SRW T K F+P LP +T
Sbjct 170 VPVEAVQAGAVVALNPAKKERRSRWS--------------TTKSFVPGMPTILPADLTED 215
Query 93 QVDRFLREQRLEELYRKLNNGILEFFDADIRPPSPPPVYDKNGGRVN 139
Q + +L + +E+ RKL + R PSP PVYD NG R+N
Sbjct 216 QRNAYLLQLEIEDATRKLRLADFGVAEGRERSPSPEPVYDANGKRLN 262
> Hs4759340
Length=623
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/76 (34%), Positives = 41/76 (53%), Gaps = 3/76 (3%)
Query 67 SRWGSET--DKPFLPPPFIDLPIGMTPLQVDRFLREQRLEELYRKLNNGILEFF-DADIR 123
SRW +T K +P +P G+T Q ++ + ++E+L RKL G L + + R
Sbjct 20 SRWNQDTMEQKTVIPGMPTVIPPGLTREQERAYIVQLQIEDLTRKLRTGDLGIPPNPEDR 79
Query 124 PPSPPPVYDKNGGRVN 139
PSP P+Y+ G R+N
Sbjct 80 SPSPEPIYNSEGKRLN 95
> SPCC962.06c
Length=587
Score = 36.2 bits (82), Expect = 0.021, Method: Composition-based stats.
Identities = 26/75 (34%), Positives = 38/75 (50%), Gaps = 5/75 (6%)
Query 69 WGSETDKPFLPPPFIDLPIG----MTPLQVDRFLREQRLEELYRKLNNGILEFFDADIRP 124
WG T + P ++L MT Q++ + RLEE+ +KL G + + R
Sbjct 73 WGHPTPIEEMLPSQMELETAVKSCMTMEQLELYSLNVRLEEITQKLRTGDVVPHHRE-RS 131
Query 125 PSPPPVYDKNGGRVN 139
PSPPP YD +G R+N
Sbjct 132 PSPPPQYDNHGRRLN 146
> Hs16418461
Length=601
Score = 35.4 bits (80), Expect = 0.031, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 28/54 (51%), Gaps = 8/54 (14%)
Query 50 EKGRKSRWEKSKKSADGSRWGS-ETDKPFLPPPFIDLPIGMTPLQVDRFLREQR 102
+KG +RWEK +K DG +W E P+ PP+ LP G+ RF E R
Sbjct 35 QKGSGARWEK-EKHEDGVKWRQLEHKGPYFAPPYEPLPDGV------RFFYEGR 81
> At3g17205
Length=873
Score = 31.2 bits (69), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 23/97 (23%), Positives = 41/97 (42%), Gaps = 15/97 (15%)
Query 22 SFLFNSFNKMSLEQLMALVPLEAAMTHKEKGRKSRWEKSKKSADGSRWGSE--------T 73
SFL+ +FN + LE++M + + ++ + W K+ + +W S
Sbjct 280 SFLYAAFNTLPLERIMTI------LAYRTELVAVLWNYMKRCHENQKWSSMPKLLAYLPG 333
Query 74 DKPFLPPPFIDLPIGMT-PLQVDRFLREQRLEELYRK 109
D P P + L +G+T L + L EE Y +
Sbjct 334 DAPGWLLPLVGLILGLTLSLMIRHMLMIVDNEEFYER 370
> YLR116w
Length=476
Score = 31.2 bits (69), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 32/118 (27%), Positives = 46/118 (38%), Gaps = 17/118 (14%)
Query 22 SFLFNSFNKMSLEQLMALVPLEAAMTHKEKGRKSRWEKSKKSADGSRWGSETDKPFLPPP 81
S F + S+E+ A VP + S W K+ +D R+ S K
Sbjct 8 SRYFENRKGSSMEEKKAKVPPNVNL--------SLWRKNTVESDVHRFNSLPSK------ 53
Query 82 FIDLPIGMTPLQVDRFLREQRLEELYRKLNNGILEFFDADIRPPSPPPVYDKNGGRVN 139
+ +T Q+ + R++E+ KL R PSPPPVYD G R N
Sbjct 54 ---ISGALTREQIYSYQVMFRIQEITIKLRTNDFVPPSRKNRSPSPPPVYDAQGKRTN 108
> 7301203
Length=1439
Score = 30.0 bits (66), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 12/34 (35%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 20 FISFLFNSFNKMSLEQLMALVPLEAAMTHKEKGR 53
F++F NSFN + + + M P + +HKE G+
Sbjct 976 FLTFATNSFNPLGIRRPMPTTPSPISQSHKEPGQ 1009
> Hs22050082
Length=1188
Score = 29.3 bits (64), Expect = 2.7, Method: Composition-based stats.
Identities = 16/67 (23%), Positives = 33/67 (49%), Gaps = 2/67 (2%)
Query 46 MTHKEKGRKSRWEKSKKSADGSRWGSETDKPFLPPPFIDLPIGMTPLQVDRF--LREQRL 103
+ H E S ++++ + D G E + PP I PI +P ++D++ L+ Q
Sbjct 946 INHVEGNINSYYDRTMQKPDKVEDGLEMCHKSISPPLIQQPITFSPDEIDKYKILQLQAQ 1005
Query 104 EELYRKL 110
+ + ++L
Sbjct 1006 QHMQKQL 1012
> At5g51300
Length=804
Score = 28.5 bits (62), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query 102 RLEELYRKLNNGI-LEFFDADIRPPSPPPVYDKNGGRVN 139
RL E+ R L +G+ L+ R PSP PVYD G R+N
Sbjct 164 RLLEISRMLQSGMPLDDRPEGQRSPSPEPVYDNMGIRIN 202
> 7296664
Length=319
Score = 28.1 bits (61), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 11/28 (39%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 81 PFIDLPIGMTPLQVDRFLREQRLEELYR 108
P +DL + P +V+R + E + EELY+
Sbjct 238 PVVDLDVASPPKRVNRDIDESQKEELYK 265
> Hs17462298
Length=283
Score = 27.7 bits (60), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 22/49 (44%), Gaps = 0/49 (0%)
Query 32 SLEQLMALVPLEAAMTHKEKGRKSRWEKSKKSADGSRWGSETDKPFLPP 80
+ Q+M LV LEA +E WE + S G ET+K L P
Sbjct 7 GINQVMKLVKLEAGKKVQESAEAPMWEMNCVSDGAQERGKETEKGDLNP 55
Lambda K H
0.323 0.140 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1534984332
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40