bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0914_orf1
Length=183
Score E
Sequences producing significant alignments: (Bits) Value
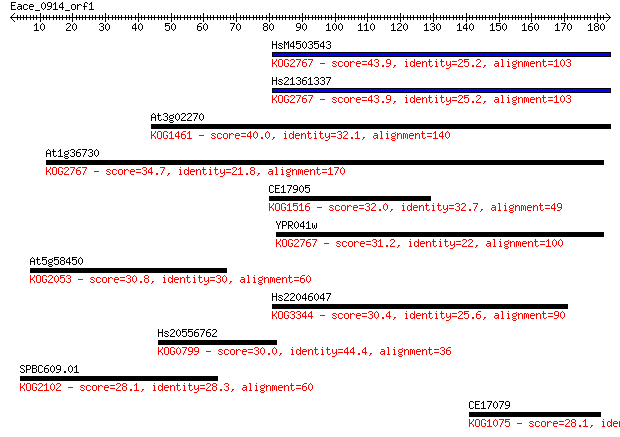
HsM4503543 43.9 2e-04
Hs21361337 43.9 2e-04
At3g02270 40.0 0.003
At1g36730 34.7 0.12
CE17905 32.0 0.68
YPR041w 31.2 1.2
At5g58450 30.8 1.5
Hs22046047 30.4 2.2
Hs20556762 30.0 2.4
SPBC609.01 28.1 8.9
CE17079 28.1 9.5
> HsM4503543
Length=431
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/107 (24%), Positives = 53/107 (49%), Gaps = 4/107 (3%)
Query 81 FVVLSALFKEGLTPEGLDQKMPFVVKCCDSSVRSSDIIYALENFCFESEETQMTGFPYLL 140
V+ LF E + + + F+ C ++ +++ LE + ++ P++L
Sbjct 278 LVLTEVLFNEKIREQIKKYRRHFLRFCHNNKKAQRYLLHGLECVVAMHQAQLISKIPHIL 337
Query 141 QRMYNAELLEAEDILNYYNADTTDPVT----LKCKTFAEPFLQWLAE 183
+ MY+A+LLE E I+++ + V+ + + AEPF++WL E
Sbjct 338 KEMYDADLLEEEVIISWSEKASKKYVSKELAKEIRVKAEPFIKWLKE 384
> Hs21361337
Length=431
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/107 (24%), Positives = 53/107 (49%), Gaps = 4/107 (3%)
Query 81 FVVLSALFKEGLTPEGLDQKMPFVVKCCDSSVRSSDIIYALENFCFESEETQMTGFPYLL 140
V+ LF E + + + F+ C ++ +++ LE + ++ P++L
Sbjct 278 LVLTEVLFNEKIREQIKKYRRHFLRFCHNNKKAQRYLLHGLECVVAMHQAQLISKIPHIL 337
Query 141 QRMYNAELLEAEDILNYYNADTTDPVT----LKCKTFAEPFLQWLAE 183
+ MY+A+LLE E I+++ + V+ + + AEPF++WL E
Sbjct 338 KEMYDADLLEEEVIISWSEKASKKYVSKELAKEIRVKAEPFIKWLKE 384
> At3g02270
Length=676
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 45/147 (30%), Positives = 65/147 (44%), Gaps = 20/147 (13%)
Query 44 NRLRNMFITTPSVTPDAFFVELRMLQVSQDFDAKCRLFVVLSALFKEGLTPEGLDQKMPF 103
N LR + + A F + L VS + L+ S++ +GL + F
Sbjct 536 NSLRLSYNMESAHCAGAIFYSMMKLAVSTPHSSINDLYRNASSIITRW---KGL---LGF 589
Query 104 VVKCCDSSVRSSDIIYALENFCFESEETQMTGFPYLLQRMYNAE--LLEAEDILNYYN-- 159
VK D + ++I LE C ES T F ++L+ MY E LL+ IL + +
Sbjct 590 YVKKSDEQI---EVISRLEEMCEESAHELGTLFAHILRYMYEEENDLLQEVAILRWSDEK 646
Query 160 --ADTTDPVTLK-CKTFAEPFLQWLAE 183
AD +D V LK C EPF+ WL E
Sbjct 647 AGADESDKVYLKQC----EPFITWLKE 669
> At1g36730
Length=439
Score = 34.7 bits (78), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 37/179 (20%), Positives = 81/179 (45%), Gaps = 19/179 (10%)
Query 12 KEKKQSGSVSEGPKLVVKEALTIRCPEIAE--VSNRLRNMFITTPSVTPDAFFVELRMLQ 69
+EKK V + P+ V E + PE A + N ++ + + S T +L+
Sbjct 261 EEKKPVAEVKKAPEQV-HENGNSKIPENAHEKLVNEIKELLSSGSSPT------QLKTAL 313
Query 70 VSQDFDAKCRLFVVLSALFK---EGLTPEGLDQK---MPFVVKCCDSSVRSS-DIIYALE 122
S + + ++ + SALF +G E + +K + ++ ++ + ++ +E
Sbjct 314 ASNSANPQEKMDALFSALFGGTGKGFAKEVIKKKKYLLALMMMQEEAGAPAQMGLLNGIE 373
Query 123 NFCFESEETQMTGFPYLLQRMYNAELLEAEDILNYYNADTTDPVTLKCKTFAEPFLQWL 181
+FC ++ +++ +Y+ ++L+ + I+ +YN LK T PF++WL
Sbjct 374 SFCMKASAEAAKEVALVIKGLYDEDILDEDVIVEWYNKGVKSSPVLKNVT---PFIEWL 429
> CE17905
Length=548
Score = 32.0 bits (71), Expect = 0.68, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 26/49 (53%), Gaps = 3/49 (6%)
Query 80 LFVVLSALFKEGLTPEGLDQKMPFVVKCCDSSVRSSDIIYALENFCFES 128
F+ L AL K PEG+ + M + K CD R+ D++ + +F F+
Sbjct 338 FFIALGALSK---NPEGIKKFMGRIFKECDYGERADDVLQMVYDFYFKG 383
> YPR041w
Length=405
Score = 31.2 bits (69), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 22/104 (21%), Positives = 46/104 (44%), Gaps = 5/104 (4%)
Query 82 VVLSALFKEGLTPEGLDQKMPFVVKCCDSSVRSSDIIYALENFCFESEETQMTGFPYLLQ 141
V+ LF E + E + + F K + + + +E F + + P +L
Sbjct 290 VLAQCLFDEDIVNE-IAEHNAFFTKILVTPEYEKNFMGGIERFLGLEHKDLIPLLPKILV 348
Query 142 RMYNAELLEAEDILNYYNADTT----DPVTLKCKTFAEPFLQWL 181
++YN +++ E+I+ + + V+ K + A+PF+ WL
Sbjct 349 QLYNNDIISEEEIMRFGTKSSKKFVPKEVSKKVRRAAKPFITWL 392
> At5g58450
Length=1035
Score = 30.8 bits (68), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 29/61 (47%), Gaps = 1/61 (1%)
Query 7 SEKKKKEKKQSGSVSEGP-KLVVKEALTIRCPEIAEVSNRLRNMFITTPSVTPDAFFVEL 65
S KKKK+ + S +S P +K+++ + C I +VSN L N + F L
Sbjct 896 SGKKKKKNQHSDQLSSSPISQAIKDSIQLLCSTIQDVSNWLLNQLNNPEDGQVEGFLTTL 955
Query 66 R 66
+
Sbjct 956 K 956
> Hs22046047
Length=155
Score = 30.4 bits (67), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 23/90 (25%), Positives = 39/90 (43%), Gaps = 2/90 (2%)
Query 81 FVVLSALFKEGLTPEGLDQKMPFVVKCCDSSVRSSDIIYALENFCFESEETQMTGFPYLL 140
VV LFKEG+ D MP ++ D +V + ++ A+++ T+ + +
Sbjct 9 IVVYELLFKEGVVVAKKDVHMPKHLELADKNVPNLHVMKAMQSLKSRGYNTEHFAWRHFY 68
Query 141 QRMYNAELLEAEDILNYYNADTTDPVTLKC 170
+ N + D L+ T PVTL C
Sbjct 69 WYLTNEGIQYLHDYLHL--PLETVPVTLCC 96
> Hs20556762
Length=374
Score = 30.0 bits (66), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 46 LRNMFITTPSVTPDAFFVELRMLQVSQDFDAKCRLF 81
L N +ITTP T +A F ++ +SQDFD LF
Sbjct 81 LENHYITTPLSTEEAAFPLAYVMTISQDFDTFEWLF 116
> SPBC609.01
Length=1157
Score = 28.1 bits (61), Expect = 8.9, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 27/60 (45%), Gaps = 0/60 (0%)
Query 4 KEKSEKKKKEKKQSGSVSEGPKLVVKEALTIRCPEIAEVSNRLRNMFITTPSVTPDAFFV 63
K +S ++ SG +GPK+V + R P IA S+++ F T D F+
Sbjct 478 KNQSSHRRNSSTSSGETGKGPKIVWFKPSDKRIPLIAISSDQVPPTFFTNNDDFKDKVFL 537
> CE17079
Length=319
Score = 28.1 bits (61), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 13/40 (32%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 141 QRMYNAELLEAEDILNYYNADTTDPVTLKCKTFAEPFLQW 180
++ NA + I N YN+ +TL KTF +P L++
Sbjct 166 KKAINASKYQLSQIFNMYNSRNPRLMTLLYKTFVKPLLEY 205
Lambda K H
0.318 0.134 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2914594326
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40