bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0908_orf1
Length=134
Score E
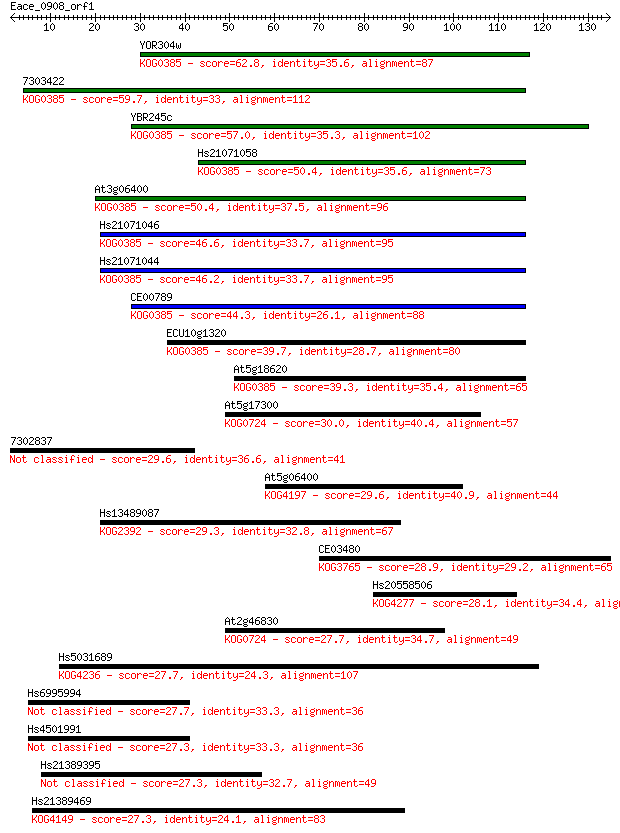
Sequences producing significant alignments: (Bits) Value
YOR304w 62.8 2e-10
7303422 59.7 1e-09
YBR245c 57.0 1e-08
Hs21071058 50.4 1e-06
At3g06400 50.4 1e-06
Hs21071046 46.6 1e-05
Hs21071044 46.2 2e-05
CE00789 44.3 7e-05
ECU10g1320 39.7 0.001
At5g18620 39.3 0.002
At5g17300 30.0 1.2
7302837 29.6 1.6
At5g06400 29.6 1.6
Hs13489087 29.3 2.2
CE03480 28.9 3.2
Hs20558506 28.1 4.3
At2g46830 27.7 6.0
Hs5031689 27.7 6.1
Hs6995994 27.7 7.0
Hs4501991 27.3 7.3
Hs21389395 27.3 8.0
Hs21389469 27.3 9.3
> YOR304w
Length=1120
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/87 (35%), Positives = 54/87 (62%), Gaps = 3/87 (3%)
Query 30 FTAEMKMQKEQLLSEGFLNWNRTEFTKFISALIHLGRDRLEEAWQLHFSSSNKTVEDLRK 89
FT E +++K++L+S+ F NWN+ +F FI+A GRD +E + S +KT E++
Sbjct 872 FTQEDELRKQELISKAFTNWNKRDFMAFINACAKYGRDDME---NIKKSIDSKTPEEVEV 928
Query 90 YADVFFKRYTEIEGGDRMMQRIERAEE 116
YA +F++R EI G ++ + +E E+
Sbjct 929 YAKIFWERLKEINGWEKYLHNVELGEK 955
> 7303422
Length=1027
Score = 59.7 bits (143), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 37/112 (33%), Positives = 60/112 (53%), Gaps = 3/112 (2%)
Query 4 PPEPEEGPSDTESALDDPDDPERPQPFTAEMKMQKEQLLSEGFLNWNRTEFTKFISALIH 63
P E G T+ ++ + +P T E +KE LLS+GF W + +F +FI A
Sbjct 755 PKNTELGSDATKVQREEQRKIDEAEPLTEEEIQEKENLLSQGFTAWTKRDFNQFIKANEK 814
Query 64 LGRDRLEEAWQLHFSSSNKTVEDLRKYADVFFKRYTEIEGGDRMMQRIERAE 115
GRD ++ + KT E++ +Y VF++R TE++ +R+M +IER E
Sbjct 815 YGRDDID---NIAKDVEGKTPEEVIEYNAVFWERCTELQDIERIMGQIERGE 863
> YBR245c
Length=1129
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 36/103 (34%), Positives = 57/103 (55%), Gaps = 4/103 (3%)
Query 28 QPFTAEMKMQKEQLLSEGFLNWNRTEFTKFISALIHLGRDRLEE-AWQLHFSSSNKTVED 86
QP T E + K SEGF NWN+ EF KFI+ GR+ ++ A +L + KT+E+
Sbjct 866 QPLTEEEEKMKADWESEGFTNWNKLEFRKFITVSGKYGRNSIQAIAREL---APGKTLEE 922
Query 87 LRKYADVFFKRYTEIEGGDRMMQRIERAEELRGAIDLQRRAIQ 129
+R YA F+ IE ++ ++ IE EE + +Q+ A++
Sbjct 923 VRAYAKAFWSNIERIEDYEKYLKIIENEEEKIKRVKMQQEALR 965
> Hs21071058
Length=1052
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 45/73 (61%), Gaps = 3/73 (4%)
Query 43 SEGFLNWNRTEFTKFISALIHLGRDRLEEAWQLHFSSSNKTVEDLRKYADVFFKRYTEIE 102
++GF NWN+ +F +FI A GRD +E + KT E++ +Y+ VF++R E++
Sbjct 839 TQGFTNWNKRDFNQFIKANEKWGRDDIE---NIAREVEGKTPEEVIEYSAVFWERCNELQ 895
Query 103 GGDRMMQRIERAE 115
+++M +IER E
Sbjct 896 DIEKIMAQIERGE 908
> At3g06400
Length=1057
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 36/96 (37%), Positives = 54/96 (56%), Gaps = 3/96 (3%)
Query 20 DPDDPERPQPFTAEMKMQKEQLLSEGFLNWNRTEFTKFISALIHLGRDRLEEAWQLHFSS 79
D ++PE P T E +KE LL EGF W+R +F F+ A GR+ ++ +
Sbjct 817 DVEEPEGGDPLTTEEVEEKEGLLEEGFSTWSRRDFNTFLRACEKYGRNDIK---SIASEM 873
Query 80 SNKTVEDLRKYADVFFKRYTEIEGGDRMMQRIERAE 115
KT E++ +YA VF +RY E+ DR+++ IER E
Sbjct 874 EGKTEEEVERYAKVFKERYKELNDYDRIIKNIERGE 909
> Hs21071046
Length=1033
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 32/111 (28%), Positives = 59/111 (53%), Gaps = 19/111 (17%)
Query 21 PDDPERPQPFTAEMKMQKE----------------QLLSEGFLNWNRTEFTKFISALIHL 64
P +P+ P P A+ + QK+ +LL++GF NW + +F +FI A
Sbjct 795 PRNPDIPNPALAQREEQKKIDGAEPLTPEETEEKEKLLTQGFTNWTKRDFNQFIKANEKY 854
Query 65 GRDRLEEAWQLHFSSSNKTVEDLRKYADVFFKRYTEIEGGDRMMQRIERAE 115
GRD ++ + K+ E++ +Y+ VF++R E++ +++M +IER E
Sbjct 855 GRDDID---NIAREVEGKSPEEVMEYSAVFWERCNELQDIEKIMAQIERGE 902
> Hs21071044
Length=1054
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 32/111 (28%), Positives = 59/111 (53%), Gaps = 19/111 (17%)
Query 21 PDDPERPQPFTAEMKMQKE----------------QLLSEGFLNWNRTEFTKFISALIHL 64
P +P+ P P A+ + QK+ +LL++GF NW + +F +FI A
Sbjct 816 PRNPDIPNPALAQREEQKKIDGAEPLTPEETEEKEKLLTQGFTNWTKRDFNQFIKANEKY 875
Query 65 GRDRLEEAWQLHFSSSNKTVEDLRKYADVFFKRYTEIEGGDRMMQRIERAE 115
GRD ++ + K+ E++ +Y+ VF++R E++ +++M +IER E
Sbjct 876 GRDDID---NIAREVEGKSPEEVMEYSAVFWERCNELQDIEKIMAQIERGE 923
> CE00789
Length=971
Score = 44.3 bits (103), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 23/88 (26%), Positives = 51/88 (57%), Gaps = 4/88 (4%)
Query 28 QPFTAEMKMQKEQLLSEGFLNWNRTEFTKFISALIHLGRDRLEEAWQLHFSSSNKTVEDL 87
+P T + + +K +LL++ +W + EF +F+ GR+ LE + + +E++
Sbjct 743 RPLTDKEQEEKAELLTQSVTDWTKREFQQFVRGNEKYGREDLESIAK----EMERPLEEI 798
Query 88 RKYADVFFKRYTEIEGGDRMMQRIERAE 115
+ YA VF++R E++ ++++ +IE+ E
Sbjct 799 QSYAKVFWERIEELQDSEKVLSQIEKGE 826
> ECU10g1320
Length=823
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 47/80 (58%), Gaps = 6/80 (7%)
Query 36 MQKEQLLSEGFLNWNRTEFTKFISALIHLGRDRLEEAWQLHFSSSNKTVEDLRKYADVFF 95
++K++L S+GF +W + +F +I A+ +GRD +E+ L K +D+ Y VF+
Sbjct 645 LRKKELQSQGF-DWTKKDFHLYIKAVEKVGRDDVEKIAMLL-----KHKDDVEAYHRVFW 698
Query 96 KRYTEIEGGDRMMQRIERAE 115
+R E+ ++++ I R++
Sbjct 699 ERVNELSDAEKIIGSINRSQ 718
> At5g18620
Length=1063
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 35/65 (53%), Gaps = 3/65 (4%)
Query 51 RTEFTKFISALIHLGRDRLEEAWQLHFSSSNKTVEDLRKYADVFFKRYTEIEGGDRMMQR 110
R +F FI A GR+ ++ + KT E++ +YA VF RY E+ DR+++
Sbjct 843 RRDFNAFIRACEKYGRNDIK---SIASEMEGKTEEEVERYAQVFQVRYKELNDYDRIIKN 899
Query 111 IERAE 115
IER E
Sbjct 900 IERGE 904
> At5g17300
Length=385
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 28/60 (46%), Gaps = 9/60 (15%)
Query 49 WNRTEFTKFISALIHLGRD--RLEEAWQLHFSSSNKTVEDLRKYADVFFKRYT-EIEGGD 105
W E KF+ AL GR R+EE H S KT +R +A FF + E GGD
Sbjct 56 WTDEEHKKFVEALKLYGRAWRRIEE----HVGS--KTAVQIRSHAQKFFSKVAREATGGD 109
> 7302837
Length=649
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 25/47 (53%), Gaps = 6/47 (12%)
Query 1 KIRPPEPEEGPSDTESALDDP------DDPERPQPFTAEMKMQKEQL 41
+I+PPEP S TE +D D P +P P ++++ ++K L
Sbjct 210 EIKPPEPNSKLSITEPIQEDNIPGTCFDTPTKPIPSSSQLSVRKSSL 256
> At5g06400
Length=1030
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 18/52 (34%), Positives = 29/52 (55%), Gaps = 8/52 (15%)
Query 58 ISALI--HLGRDRLEEAWQLHFSSSNKTVEDLRKYADVFFK------RYTEI 101
I+A++ HLG++R+ EAW++ S K ++ K +F K RY EI
Sbjct 471 ITAVVAGHLGQNRVAEAWKVFSSMEEKGIKPTWKSYSIFVKELCRSSRYDEI 522
> Hs13489087
Length=379
Score = 29.3 bits (64), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 33/67 (49%), Gaps = 3/67 (4%)
Query 21 PDDPERPQPFTAEMKMQKEQLLSEGFLNWNRTEFTKFISALIHLGRDRLEEAWQLHFSSS 80
PDD E + +K +EQL E W + E FI + L R +LEE++ L+ +
Sbjct 233 PDDIEDE---STGLKKIEEQLTLEKLHEWTKPENLDFIEVNVSLPRFKLEESYTLNSDLA 289
Query 81 NKTVEDL 87
V+DL
Sbjct 290 RLGVQDL 296
> CE03480
Length=622
Score = 28.9 bits (63), Expect = 3.2, Method: Composition-based stats.
Identities = 19/65 (29%), Positives = 31/65 (47%), Gaps = 2/65 (3%)
Query 70 EEAWQLHFSSSNKTVEDLRKYADVFFKRYTEIEGGDRMMQRIERAEELRGAIDLQRRAIQ 129
E LHF+S NKTV + D K + E++G D ++R R+ + D+ +
Sbjct 263 ETPLALHFNSQNKTVGKNYAFFDKIRKAFDEMDGSD--LKRRRRSFKGNNQKDICHEYLP 320
Query 130 ATSFR 134
+FR
Sbjct 321 LDNFR 325
> Hs20558506
Length=454
Score = 28.1 bits (61), Expect = 4.3, Method: Composition-based stats.
Identities = 11/32 (34%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 82 KTVEDLRKYADVFFKRYTEIEGGDRMMQRIER 113
K VED+ ++ + E +GGD ++QR++R
Sbjct 327 KNVEDMVQFINNILDGTVEAQGGDSILQRLKR 358
> At2g46830
Length=608
Score = 27.7 bits (60), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 23/50 (46%), Gaps = 6/50 (12%)
Query 49 WNRTEFTKFISALIHLGRDRLEEAWQ-LHFSSSNKTVEDLRKYADVFFKR 97
W E +FI AL GR AWQ + + KT +R +A FF +
Sbjct 27 WTEEEHNRFIEALRLYGR-----AWQKIEEHVATKTAVQIRSHAQKFFSK 71
> Hs5031689
Length=890
Score = 27.7 bits (60), Expect = 6.1, Method: Composition-based stats.
Identities = 26/119 (21%), Positives = 52/119 (43%), Gaps = 14/119 (11%)
Query 12 SDTESALDDPDDPERPQ-------PFTAEMKMQKEQLLSEGFLNWNRTEFTKFISALIHL 64
SD+ LDD ++P P+ P +++ +E + + N + + ++ H
Sbjct 351 SDSSRGLDDTEEPSPPEDKMFFLDPSDLDVERDEEAVKTISPSTSNNIPLMRVVQSIKHT 410
Query 65 GRDR---LEEAWQLHFSSSNKTVEDLRKYADVFFKRYT--EIEGGDRMMQRIERAEELR 118
R ++E W +H++S + + R Y + K T + E G + + I +E LR
Sbjct 411 KRKSSTMVKEGWMVHYTSRDNLRK--RHYWRLDSKCLTLFQNESGSKYYKEIPLSEILR 467
> Hs6995994
Length=2415
Score = 27.7 bits (60), Expect = 7.0, Method: Composition-based stats.
Identities = 12/36 (33%), Positives = 18/36 (50%), Gaps = 0/36 (0%)
Query 5 PEPEEGPSDTESALDDPDDPERPQPFTAEMKMQKEQ 40
P E PS+ SA ++P P P+P E+ E+
Sbjct 824 PSKEPSPSEEPSASEEPYTPSPPEPSWTELPSSGEE 859
> Hs4501991
Length=2316
Score = 27.3 bits (59), Expect = 7.3, Method: Composition-based stats.
Identities = 12/36 (33%), Positives = 18/36 (50%), Gaps = 0/36 (0%)
Query 5 PEPEEGPSDTESALDDPDDPERPQPFTAEMKMQKEQ 40
P E PS+ SA ++P P P+P E+ E+
Sbjct 824 PSKEPSPSEEPSASEEPYTPSPPEPSWTELPSSGEE 859
> Hs21389395
Length=265
Score = 27.3 bits (59), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 16/52 (30%), Positives = 27/52 (51%), Gaps = 4/52 (7%)
Query 8 EEGPSDTESALDDPDDPE---RPQPFTAEMKMQKEQLLSEGFLNWNRTEFTK 56
++GP +++ + D DPE RP PF + + + +L EG N R F +
Sbjct 125 KDGPKKSDTDIKDAVDPESTQRPNPFRRQ-SIVLDPMLQEGTFNSQRATFIR 175
> Hs21389469
Length=155
Score = 27.3 bits (59), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 20/83 (24%), Positives = 32/83 (38%), Gaps = 17/83 (20%)
Query 6 EPEEGPSDTESALDDPDDPERPQPFTAEMKMQKEQLLSEGFLNWNRTEFTKFISALIHLG 65
E E PS E DDP+DP + +L G +NWN L +
Sbjct 31 EDHETPSSAELVADDPNDPYE----------EHGLILPNGNINWN-------CPCLGGMA 73
Query 66 RDRLEEAWQLHFSSSNKTVEDLR 88
E ++ FS + + E+++
Sbjct 74 SGPCGEQFKSAFSCFHYSTEEIK 96
Lambda K H
0.316 0.134 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1393086308
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40