bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0887_orf4
Length=190
Score E
Sequences producing significant alignments: (Bits) Value
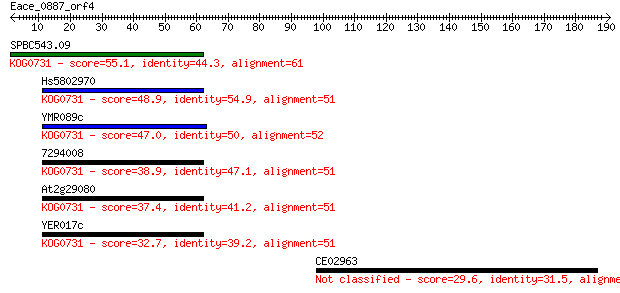
SPBC543.09 55.1 8e-08
Hs5802970 48.9 6e-06
YMR089c 47.0 3e-05
7294008 38.9 0.006
At2g29080 37.4 0.017
YER017c 32.7 0.41
CE02963 29.6 4.2
> SPBC543.09
Length=773
Score = 55.1 bits (131), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 27/72 (37%), Positives = 44/72 (61%), Gaps = 14/72 (19%)
Query 1 RQSPQDPYQLFSDATAQLVDDEVRNLISNQYERVKALLKEKE-----------KKETLTF 49
R++ Q P FS+ATAQ++D+E+R L+ + YER K LL E + +KE +T+
Sbjct 669 RETVQKP---FSEATAQMIDEEIRKLVKHAYERTKKLLLEHKQGLENIAQRLLQKEVITY 725
Query 50 ADLQDCLGTRPF 61
+++ LG RP+
Sbjct 726 NEVETILGPRPY 737
> Hs5802970
Length=797
Score = 48.9 bits (115), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 39/62 (62%), Gaps = 11/62 (17%)
Query 11 FSDATAQLVDDEVRNLISNQYERVKALLKEKE-----------KKETLTFADLQDCLGTR 59
+S+ATA+L+DDEVR LI++ Y+R ALL EK+ +KE L D+ + LG R
Sbjct 689 YSEATARLIDDEVRILINDAYKRTVALLTEKKADVEKVALLLLEKEVLDKNDMVELLGPR 748
Query 60 PF 61
PF
Sbjct 749 PF 750
> YMR089c
Length=825
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 26/63 (41%), Positives = 33/63 (52%), Gaps = 11/63 (17%)
Query 11 FSDATAQLVDDEVRNLISNQYERVKALLKEKE-----------KKETLTFADLQDCLGTR 59
FSD T ++D EV ++ ++R LLKEK KKE LT D+ D LG R
Sbjct 727 FSDETGDIIDSEVYRIVQECHDRCTKLLKEKAEDVEKIAQVLLKKEVLTREDMIDLLGKR 786
Query 60 PFP 62
PFP
Sbjct 787 PFP 789
> 7294008
Length=793
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 24/62 (38%), Positives = 36/62 (58%), Gaps = 11/62 (17%)
Query 11 FSDATAQLVDDEVRNLISNQYERVKALL-KEKE----------KKETLTFADLQDCLGTR 59
+S+ TAQL+D+EVR++I +E +LL K KE + E L+ D+ + LG R
Sbjct 674 YSEDTAQLIDNEVRSIIKCAHEATTSLLTKHKENVQKVAERLLQNEVLSRDDMIELLGPR 733
Query 60 PF 61
PF
Sbjct 734 PF 735
> At2g29080
Length=807
Score = 37.4 bits (85), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 33/62 (53%), Gaps = 11/62 (17%)
Query 11 FSDATAQLVDDEVRNLISNQYERVKALLKEKE-----------KKETLTFADLQDCLGTR 59
+S+ T ++D+EVR+ ++ YER L++E + +KE L DL LG R
Sbjct 699 YSNKTGAIIDEEVRDWVAKAYERTVELVEEHKVKVAEIAELLLEKEVLHQDDLLKILGER 758
Query 60 PF 61
PF
Sbjct 759 PF 760
> YER017c
Length=761
Score = 32.7 bits (73), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 32/62 (51%), Gaps = 11/62 (17%)
Query 11 FSDATAQLVDDEVRNLISNQYERVKALL-----------KEKEKKETLTFADLQDCLGTR 59
FS+ TA+ +D EV++++ + + LL KE +KE +T D+ LG R
Sbjct 673 FSNKTARTIDLEVKSIVDDAHRACTELLTKNLDKVDLVAKELLRKEAITREDMIRLLGPR 732
Query 60 PF 61
PF
Sbjct 733 PF 734
> CE02963
Length=1225
Score = 29.6 bits (65), Expect = 4.2, Method: Composition-based stats.
Identities = 28/97 (28%), Positives = 41/97 (42%), Gaps = 10/97 (10%)
Query 98 IPT--AISGDRNVRDEKGSTSDKDAAADGHSMQQRKQSIKDTEGSNGDDKEPTDAGKNGG 155
IPT IS D + E ++K Q K++ + + G +E ++ +
Sbjct 511 IPTQRTISCDDDGYSESTRNNNKAPQVFSPDKQDAKKNCQVISENFGKSEEQSEGDETSN 570
Query 156 KGKKKRNGKGNAADDDDDDNDDD------DDDRNGNN 186
KG K N GN DD D +N D DD+ NG N
Sbjct 571 KGSKDEN--GNMLDDVDQENTSDNEISFPDDEANGRN 605
Lambda K H
0.305 0.128 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3166472848
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40