bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0846_orf1
Length=118
Score E
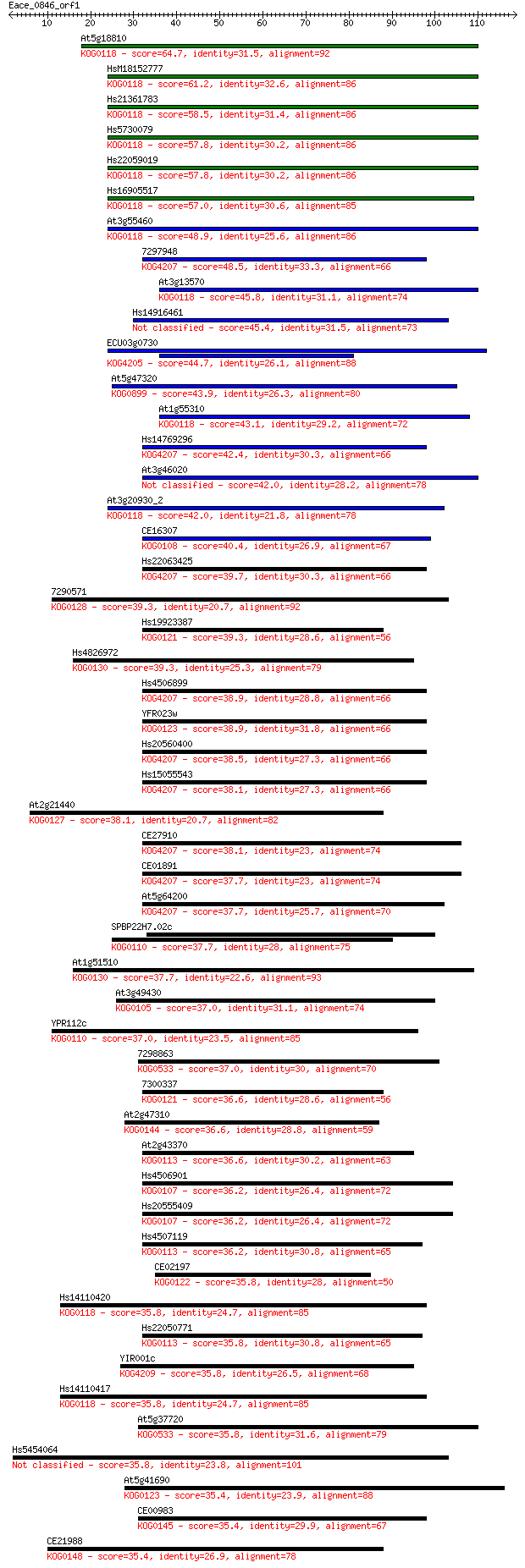
Sequences producing significant alignments: (Bits) Value
At5g18810 64.7 4e-11
HsM18152777 61.2 4e-10
Hs21361783 58.5 3e-09
Hs5730079 57.8 4e-09
Hs22059019 57.8 4e-09
Hs16905517 57.0 9e-09
At3g55460 48.9 2e-06
7297948 48.5 3e-06
At3g13570 45.8 2e-05
Hs14916461 45.4 2e-05
ECU03g0730 44.7 4e-05
At5g47320 43.9 7e-05
At1g55310 43.1 1e-04
Hs14769296 42.4 2e-04
At3g46020 42.0 2e-04
At3g20930_2 42.0 3e-04
CE16307 40.4 8e-04
Hs22063425 39.7 0.001
7290571 39.3 0.002
Hs19923387 39.3 0.002
Hs4826972 39.3 0.002
Hs4506899 38.9 0.002
YFR023w 38.9 0.003
Hs20560400 38.5 0.003
Hs15055543 38.1 0.003
At2g21440 38.1 0.003
CE27910 38.1 0.004
CE01891 37.7 0.005
At5g64200 37.7 0.006
SPBP22H7.02c 37.7 0.006
At1g51510 37.7 0.006
At3g49430 37.0 0.008
YPR112c 37.0 0.009
7298863 37.0 0.009
7300337 36.6 0.010
At2g47310 36.6 0.012
At2g43370 36.6 0.013
Hs4506901 36.2 0.014
Hs20555409 36.2 0.014
Hs4507119 36.2 0.015
CE02197 35.8 0.018
Hs14110420 35.8 0.018
Hs22050771 35.8 0.018
YIR001c 35.8 0.019
Hs14110417 35.8 0.019
At5g37720 35.8 0.020
Hs5454064 35.8 0.021
At5g41690 35.4 0.022
CE00983 35.4 0.024
CE21988 35.4 0.025
> At5g18810
Length=147
Score = 64.7 bits (156), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 29/92 (31%), Positives = 50/92 (54%), Gaps = 0/92 (0%)
Query 18 MRKFPYMLRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGER 77
+R P R + P P+D+R + G +K++YLP ++ PR + F Y D
Sbjct 51 IRNLPLDARSLRPSLLPNDLRDSFERFGPLKDIYLPRNYYTGEPRGFGFVKYRYAEDAAE 110
Query 78 AMKELNNQMLMGFRVTVQMANSDPKTPEQMRT 109
AMK +N++++ G + + A + KTP++MRT
Sbjct 111 AMKRMNHKVIGGREIAIVFAEENRKTPQEMRT 142
> HsM18152777
Length=261
Score = 61.2 bits (147), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 28/86 (32%), Positives = 47/86 (54%), Gaps = 0/86 (0%)
Query 24 MLRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELN 83
+R VA T P D+R + G + ++Y+P+ R PR + + +E D E A+ LN
Sbjct 13 FIRNVADATRPEDLRREFGRYGPIVDVYIPLDFYTRRPRGFAYVQFEDVRDAEDALYNLN 72
Query 84 NQMLMGFRVTVQMANSDPKTPEQMRT 109
+ + G ++ +Q A D KTP QM++
Sbjct 73 RKWVCGRQIEIQFAQGDRKTPGQMKS 98
> Hs21361783
Length=261
Score = 58.5 bits (140), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 46/86 (53%), Gaps = 0/86 (0%)
Query 24 MLRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELN 83
+R VA T P D+R + G + ++Y+P+ R PR + + +E E A+ LN
Sbjct 13 FIRNVADATRPEDLRREFGRYGPIVDVYIPLDFYTRRPRGFAYVQFEDVRGAEDALYNLN 72
Query 84 NQMLMGFRVTVQMANSDPKTPEQMRT 109
+ + G ++ +Q A D KTP QM++
Sbjct 73 RKWVCGRQIEIQFAQGDRKTPGQMKS 98
> Hs5730079
Length=183
Score = 57.8 bits (138), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 45/86 (52%), Gaps = 0/86 (0%)
Query 24 MLRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELN 83
+R VA T D+R + G + ++Y+P+ R PR + + +E D E A+ L+
Sbjct 13 FVRNVADDTRSEDLRREFGRYGPIVDVYVPLDFYTRRPRGFAYVQFEDVRDAEDALHNLD 72
Query 84 NQMLMGFRVTVQMANSDPKTPEQMRT 109
+ + G ++ +Q A D KTP QM+
Sbjct 73 RKWICGRQIEIQFAQGDRKTPNQMKA 98
> Hs22059019
Length=182
Score = 57.8 bits (138), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 45/86 (52%), Gaps = 0/86 (0%)
Query 24 MLRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELN 83
+R VA T D+R + G + ++Y+P+ R PR + + +E D E A+ L+
Sbjct 13 FVRNVADDTRSEDLRREFGRYGPIVDVYVPLDFYTRRPRGFAYVQFEDVRDAEDALHNLD 72
Query 84 NQMLMGFRVTVQMANSDPKTPEQMRT 109
+ + G ++ +Q A D KTP QM+
Sbjct 73 RKWICGRQIEIQFAQGDRKTPNQMKA 98
> Hs16905517
Length=262
Score = 57.0 bits (136), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 26/85 (30%), Positives = 45/85 (52%), Gaps = 0/85 (0%)
Query 24 MLRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELN 83
+R VA T D+R + G + ++Y+P+ R PR + + +E D E A+ L+
Sbjct 13 FVRNVADDTRSEDLRREFGRYGPIVDVYVPLDFYTRRPRGFAYVQFEDVRDAEDALHNLD 72
Query 84 NQMLMGFRVTVQMANSDPKTPEQMR 108
+ + G ++ +Q A D KTP QM+
Sbjct 73 RKWICGRQIEIQFAQGDRKTPNQMK 97
> At3g55460
Length=309
Score = 48.9 bits (115), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 22/86 (25%), Positives = 43/86 (50%), Gaps = 0/86 (0%)
Query 24 MLRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELN 83
++R + P ++R + G V+++Y+P + PR + F ++ D A + +N
Sbjct 50 LVRNIPLDCRPEELREPFERFGPVRDVYIPRDYYSGQPRGFAFVEFVDAYDAGEAQRSMN 109
Query 84 NQMLMGFRVTVQMANSDPKTPEQMRT 109
+ G +TV +A+ K PE+MR
Sbjct 110 RRSFAGREITVVVASESRKRPEEMRV 135
> 7297948
Length=195
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 37/66 (56%), Gaps = 0/66 (0%)
Query 32 TTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGFR 91
TTP D+R + G+V ++Y+P R R + F + + D E A++ ++ +ML G
Sbjct 34 TTPEDLRRVFERCGEVGDIYIPRDRYTRESRGFAFVRFYDKRDAEDALEAMDGRMLDGRE 93
Query 92 VTVQMA 97
+ VQMA
Sbjct 94 LRVQMA 99
> At3g13570
Length=262
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 39/74 (52%), Gaps = 0/74 (0%)
Query 36 DVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGFRVTVQ 95
D+R + G VK++YLP + PR + F + +D A +++ +L+G +TV
Sbjct 52 DLRRPFEQFGPVKDIYLPRDYYTGDPRGFGFIQFMDPADAAEAKHQMDGYLLLGRELTVV 111
Query 96 MANSDPKTPEQMRT 109
A + K P +MRT
Sbjct 112 FAEENRKKPTEMRT 125
> Hs14916461
Length=190
Score = 45.4 bits (106), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 41/76 (53%), Gaps = 3/76 (3%)
Query 30 PGTTPSDVRSFLSKIGQVKEMYLPVHH---LERTPRSYCFFDYERESDGERAMKELNNQM 86
P T + L K G+VK+ H LE PR YCF ++E + + E+A++ LN ++
Sbjct 34 PKITEYHLLKLLQKFGKVKQFDFLFHKSGALEGQPRGYCFVNFETKQEAEQAIQCLNGKL 93
Query 87 LMGFRVTVQMANSDPK 102
+ ++ V+ A++ K
Sbjct 94 ALSKKLVVRWAHAQVK 109
> ECU03g0730
Length=293
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/88 (26%), Positives = 52/88 (59%), Gaps = 3/88 (3%)
Query 24 MLRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELN 83
++G++ T D++S + KIG+V + +P+ + + + + + ++ +E D ++A+K L+
Sbjct 90 FIKGISYDLTEYDLKSEMEKIGKVARVGIPMTNDHKRNKGFGYVEFCKEEDVKKALK-LD 148
Query 84 NQMLMGFRVTVQMANSDPKTPEQMRTIY 111
+ +G V V MA+ P+ ++ TIY
Sbjct 149 GTVFLGREVVVNMAH--PRANKERHTIY 174
Score = 28.5 bits (62), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 22/45 (48%), Gaps = 0/45 (0%)
Query 36 DVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMK 80
D++ + +G+V M LP + Y F D+ + D ER +K
Sbjct 186 DLKKYFEGMGEVVGMSLPYDRDNNRLKGYGFVDFGNKEDYERVLK 230
> At5g47320
Length=244
Score = 43.9 bits (102), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 21/80 (26%), Positives = 36/80 (45%), Gaps = 0/80 (0%)
Query 25 LRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNN 84
+ G++PGT ++ S V E + + + R Y F ++ E A+ +N
Sbjct 67 IGGLSPGTDEHSLKDAFSSFNGVTEARVMTNKVTGRSRGYGFVNFISEDSANSAISAMNG 126
Query 85 QMLMGFRVTVQMANSDPKTP 104
Q L GF ++V +A P P
Sbjct 127 QELNGFNISVNVAKDWPSLP 146
> At1g55310
Length=220
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 36/72 (50%), Gaps = 0/72 (0%)
Query 36 DVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGFRVTVQ 95
D+R + G VK++YLP + PR + F + +D A ++ +L+G +TV
Sbjct 51 DLRKSFEQFGPVKDIYLPRDYYTGDPRGFGFVQFMDPADAADAKHHMDGYLLLGRELTVV 110
Query 96 MANSDPKTPEQM 107
A + K P +M
Sbjct 111 FAEENRKKPTEM 122
> Hs14769296
Length=221
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 34/66 (51%), Gaps = 0/66 (0%)
Query 32 TTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGFR 91
T+P +R K G+V ++Y+P + R + F + + D E AM ++ +L G
Sbjct 25 TSPDTLRRVFEKYGRVGDVYIPRDRYTKESRGFAFVRFHDKRDAEDAMDAMDGAVLDGRE 84
Query 92 VTVQMA 97
+ VQMA
Sbjct 85 LRVQMA 90
> At3g46020
Length=102
Score = 42.0 bits (97), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 40/78 (51%), Gaps = 4/78 (5%)
Query 32 TTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGFR 91
TT +R S GQ+KE L + P+ + F ++ E D +A+K L+ +++ G
Sbjct 18 TTDQSLRQLFSPFGQIKEARLIRDSETQRPKGFGFITFDSEDDARKALKSLDGKIVDGRL 77
Query 92 VTVQMANSDPKTPEQMRT 109
+ V++A K E++R
Sbjct 78 IFVEVA----KNAEEVRA 91
> At3g20930_2
Length=92
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 17/78 (21%), Positives = 41/78 (52%), Gaps = 0/78 (0%)
Query 24 MLRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELN 83
+ G++ T+ +R+ G++ E+ + + + + + Y F +Y E A+KE+N
Sbjct 3 FITGLSFYTSEKTLRAAFEGFGELVEVKIIMDKISKRSKGYAFLEYTTEEAAGTALKEMN 62
Query 84 NQMLMGFRVTVQMANSDP 101
+++ G+ + V +A + P
Sbjct 63 GKIINGWMIVVDVAKTKP 80
> CE16307
Length=83
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 18/67 (26%), Positives = 34/67 (50%), Gaps = 0/67 (0%)
Query 32 TTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGFR 91
TT ++ +F S IGQ+ + + PR + F ++ E +RA++++N G
Sbjct 16 TTEEELGNFFSSIGQINNVRIVCDRETGRPRGFAFIEFAEEGSAQRAVEQMNGAEFNGRP 75
Query 92 VTVQMAN 98
+ V +AN
Sbjct 76 LRVNLAN 82
> Hs22063425
Length=293
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 34/66 (51%), Gaps = 0/66 (0%)
Query 32 TTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGFR 91
T+P +R K G+V ++Y+P + PR + F + SD + A ++ +L G
Sbjct 25 TSPDCLRRVFEKYGRVGDVYIPREPHTKAPRGFAFVRFHYRSDAQDAEAAMDWAVLDGRE 84
Query 92 VTVQMA 97
+ VQMA
Sbjct 85 LRVQMA 90
> 7290571
Length=941
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/92 (20%), Positives = 44/92 (47%), Gaps = 1/92 (1%)
Query 11 NAKTARTMRKFPYMLRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYE 70
N K + M +R + P + ++ S G +K++ L VH L + + + ++E
Sbjct 714 NFKYSPNMEINKIFVRNLHPACSKEELHELFSPFGTIKDVRL-VHKLNKQFKGIAYVEFE 772
Query 71 RESDGERAMKELNNQMLMGFRVTVQMANSDPK 102
+ + +RA+ + + G ++V ++N P+
Sbjct 773 KPGEAQRAVAGRDGCLFKGMNISVAISNPPPR 804
> Hs19923387
Length=156
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 16/56 (28%), Positives = 30/56 (53%), Gaps = 0/56 (0%)
Query 32 TTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQML 87
TT + SK G +K++ + + +++T +CF +Y +D E AM+ +N L
Sbjct 51 TTEEQIYELFSKSGDIKKIIMGLDKMKKTACGFCFVEYYSRADAENAMRYINGTRL 106
> Hs4826972
Length=174
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/79 (25%), Positives = 39/79 (49%), Gaps = 0/79 (0%)
Query 16 RTMRKFPYMLRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDG 75
R++ + + GV T D+ ++ G++K ++L + + Y +YE +
Sbjct 68 RSVEGWILFVTGVHEEATEEDIHDKFAEYGEIKNIHLNLDRRTGYLKGYTLVEYETYKEA 127
Query 76 ERAMKELNNQMLMGFRVTV 94
+ AM+ LN Q LMG ++V
Sbjct 128 QAAMEGLNGQDLMGQPISV 146
> Hs4506899
Length=221
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 33/66 (50%), Gaps = 0/66 (0%)
Query 32 TTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGFR 91
T+P +R K +V ++Y+P + R + F + + D E AM ++ +L G
Sbjct 25 TSPDTLRRVFEKYRRVGDVYIPRDRYTKESRGFAFVRFHDKRDAEDAMDAMDGAVLDGRE 84
Query 92 VTVQMA 97
+ VQMA
Sbjct 85 LRVQMA 90
> YFR023w
Length=611
Score = 38.9 bits (89), Expect = 0.003, Method: Composition-based stats.
Identities = 21/66 (31%), Positives = 35/66 (53%), Gaps = 2/66 (3%)
Query 32 TTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGFR 91
TT D+ +F S++G +K +YL + + + F Y+ SD E+A+K NN G +
Sbjct 314 TTRDDILNFFSEVGPIKSIYLS--NATKVKYLWAFVTYKNSSDSEKAIKRYNNFYFRGKK 371
Query 92 VTVQMA 97
+ V A
Sbjct 372 LLVTRA 377
> Hs20560400
Length=234
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 18/66 (27%), Positives = 34/66 (51%), Gaps = 0/66 (0%)
Query 32 TTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGFR 91
T+P +R K G+V ++Y+P+ + PR + F + SD + A ++ +L
Sbjct 25 TSPDSLRRVFEKYGRVGDVYIPLEPHTKAPRGFAFVRFHDRSDAQDAEAAMDGAVLDERE 84
Query 92 VTVQMA 97
+ V+MA
Sbjct 85 LRVRMA 90
> Hs15055543
Length=282
Score = 38.1 bits (87), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 18/66 (27%), Positives = 32/66 (48%), Gaps = 0/66 (0%)
Query 32 TTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGFR 91
T+P +R K G+V ++Y+P + PR + F + D + A ++ L G
Sbjct 25 TSPDSLRRVFEKYGRVGDVYIPREPHTKAPRGFAFVRFHDRRDAQDAEAAMDGAELDGRE 84
Query 92 VTVQMA 97
+ VQ+A
Sbjct 85 LRVQVA 90
> At2g21440
Length=1003
Score = 38.1 bits (87), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 17/82 (20%), Positives = 38/82 (46%), Gaps = 0/82 (0%)
Query 6 LWLVENAKTARTMRKFPYMLRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYC 65
+W + +K+ ++R + PSD++ S +G V ++++P + P+ +
Sbjct 316 IWARQLGGEGSKAQKWKLIIRNLPFQAKPSDIKVVFSAVGFVWDVFIPKNFETGLPKGFA 375
Query 66 FFDYERESDGERAMKELNNQML 87
F + + D A+K+ N M
Sbjct 376 FVKFTCKKDAANAIKKFNGHMF 397
> CE27910
Length=126
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 17/74 (22%), Positives = 36/74 (48%), Gaps = 0/74 (0%)
Query 32 TTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGFR 91
TTP+D+R + G + ++++P R + + F + D E A+ + +++ G
Sbjct 30 TTPNDLRRTFERYGDIGDVHIPRDKYSRQSKGFGFVRFYERRDAEHALDRTDGKLVDGRE 89
Query 92 VTVQMANSDPKTPE 105
+ V +A D + E
Sbjct 90 LRVTLAKYDRPSDE 103
> CE01891
Length=196
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 17/74 (22%), Positives = 36/74 (48%), Gaps = 0/74 (0%)
Query 32 TTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGFR 91
TTP+D+R + G + ++++P R + + F + D E A+ + +++ G
Sbjct 30 TTPNDLRRTFERYGDIGDVHIPRDKYSRQSKGFGFVRFYERRDAEHALDRTDGKLVDGRE 89
Query 92 VTVQMANSDPKTPE 105
+ V +A D + E
Sbjct 90 LRVTLAKYDRPSDE 103
> At5g64200
Length=303
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 18/70 (25%), Positives = 37/70 (52%), Gaps = 0/70 (0%)
Query 32 TTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGFR 91
TT D+ +K G+V ++++P R + F Y+ + + +A++ L+ +++ G
Sbjct 27 TTADDLYPLFAKYGKVVDVFIPRDRRTGDSRGFAFVRYKYKDEAHKAVERLDGRVVDGRE 86
Query 92 VTVQMANSDP 101
+TVQ A P
Sbjct 87 ITVQFAKYGP 96
> SPBP22H7.02c
Length=833
Score = 37.7 bits (86), Expect = 0.006, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 36/67 (53%), Gaps = 1/67 (1%)
Query 33 TPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGFRV 92
T DV+S L GQ++ + +P +R+ R + F ++ + AM+ L N L+G +
Sbjct 733 TKKDVQSLLGAYGQLRSVRVP-KKFDRSARGFAFAEFVTAREAANAMRALKNTHLLGRHL 791
Query 93 TVQMANS 99
+Q A++
Sbjct 792 VLQYASN 798
Score = 37.0 bits (84), Expect = 0.009, Method: Composition-based stats.
Identities = 15/65 (23%), Positives = 31/65 (47%), Gaps = 0/65 (0%)
Query 25 LRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNN 84
LR + D++S GQ++++++P+ P+ + + D+ D RA EL+
Sbjct 327 LRNLTYSCAEDDLKSLFGPFGQLEQVHMPIDKKTNNPKGFAYIDFHDADDAVRAYLELDA 386
Query 85 QMLMG 89
+ G
Sbjct 387 KPFQG 391
> At1g51510
Length=191
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 21/94 (22%), Positives = 43/94 (45%), Gaps = 1/94 (1%)
Query 16 RTMRKFPYMLRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDG 75
R++ + ++ GV T D+ + G++K + L + + Y +YE++ +
Sbjct 90 RSVEGWIILVSGVHEETQEEDITNAFGDFGEIKNLNLNLDRRSGYVKGYALIEYEKKEEA 149
Query 76 ERAMKELNNQMLMGFRVTVQMA-NSDPKTPEQMR 108
+ A+ +N L+ V+V A +S P E R
Sbjct 150 QSAISAMNGAELLTQNVSVDWAFSSGPSGGESYR 183
> At3g49430
Length=243
Score = 37.0 bits (84), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 37/79 (46%), Gaps = 8/79 (10%)
Query 26 RGVAPGTTPSDVR-----SFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMK 80
R + G P D+R K G++ ++ L V P YCF ++E D E A+K
Sbjct 7 RSIYVGNLPGDIREHEIEDIFYKYGRIVDIELKV---PPRPPCYCFVEFEHSRDAEDAIK 63
Query 81 ELNNQMLMGFRVTVQMANS 99
+ L G R+ V++A+
Sbjct 64 GRDGYNLDGCRLRVELAHG 82
> YPR112c
Length=887
Score = 37.0 bits (84), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 20/85 (23%), Positives = 41/85 (48%), Gaps = 1/85 (1%)
Query 11 NAKTARTMRKFPYMLRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYE 70
N KT + +++ + T DV + GQ+K + +P +++ R + F ++
Sbjct 753 NTKTKSNKKSGKIIVKNLPFEATRKDVFELFNSFGQLKSVRVP-KKFDKSARGFAFVEFL 811
Query 71 RESDGERAMKELNNQMLMGFRVTVQ 95
+ E AM +L+ L+G R+ +Q
Sbjct 812 LPKEAENAMDQLHGVHLLGRRLVMQ 836
> 7298863
Length=266
Score = 37.0 bits (84), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 40/72 (55%), Gaps = 5/72 (6%)
Query 31 GTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFD--YERESDGERAMKELNNQMLM 88
G + +D++ + G +K+ + H +R+ RS D +ER +D +A+K+ + L
Sbjct 119 GVSNTDIKELFNDFGPIKKAAV---HYDRSGRSLGTADVIFERRADALKAIKQYHGVPLD 175
Query 89 GFRVTVQMANSD 100
G +T+Q+A SD
Sbjct 176 GRPMTIQLAVSD 187
> 7300337
Length=154
Score = 36.6 bits (83), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 16/56 (28%), Positives = 29/56 (51%), Gaps = 0/56 (0%)
Query 32 TTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQML 87
TT + S+ G V+ + + + ++TP +CF +Y S+ E AM+ +N L
Sbjct 41 TTEEQIHELFSRCGDVRVIVMGLDKYKKTPCGFCFVEYYVRSEAEAAMRFVNGTRL 96
> At2g47310
Length=324
Score = 36.6 bits (83), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 27/59 (45%), Gaps = 0/59 (0%)
Query 28 VAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQM 86
++ T D+R K G V E+ LP + +YCF Y++ +G A+ L Q
Sbjct 117 ISKTATEYDIRQVFEKYGNVTEIILPKDKMTGERAAYCFIKYKKVEEGNAAIAALTEQF 175
> At2g43370
Length=292
Score = 36.6 bits (83), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 34/63 (53%), Gaps = 0/63 (0%)
Query 32 TTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGFR 91
TT +R +SK G++K + L H + R Y F +YE E + RA ++ ++ ++ G
Sbjct 69 TTEDTLREVMSKYGRIKNLRLVRHIVTGASRGYGFVEYETEKEMLRAYEDAHHSLIDGRE 128
Query 92 VTV 94
+ V
Sbjct 129 IIV 131
> Hs4506901
Length=164
Score = 36.2 bits (82), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 19/83 (22%), Positives = 39/83 (46%), Gaps = 11/83 (13%)
Query 32 TTPSDVRSFLSKIGQVKE---------MYLPVHHL--ERTPRSYCFFDYERESDGERAMK 80
+ P D + ++ +G Y P+ + R P + F ++E D A++
Sbjct 5 SCPLDCKVYVGNLGNNGNKTELERAFGYYGPLRSVWVARNPPGFAFVEFEDPRDAADAVR 64
Query 81 ELNNQMLMGFRVTVQMANSDPKT 103
EL+ + L G RV V+++N + ++
Sbjct 65 ELDGRTLCGCRVRVELSNGEKRS 87
> Hs20555409
Length=124
Score = 36.2 bits (82), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 19/83 (22%), Positives = 39/83 (46%), Gaps = 11/83 (13%)
Query 32 TTPSDVRSFLSKIGQVKE---------MYLPVHHL--ERTPRSYCFFDYERESDGERAMK 80
+ P D + ++ +G Y P+ + R P + F ++E D A++
Sbjct 5 SCPLDCKVYVGNLGNNGNKTELERAFGYYGPLRSVWVARNPPGFAFVEFEDPRDAADAVR 64
Query 81 ELNNQMLMGFRVTVQMANSDPKT 103
EL+ + L G RV V+++N + ++
Sbjct 65 ELDGRTLCGCRVRVELSNGEKRS 87
> Hs4507119
Length=614
Score = 36.2 bits (82), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 30/65 (46%), Gaps = 0/65 (0%)
Query 32 TTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGFR 91
TT S +R G +K +++ PR Y F +YE E D A K + + + G R
Sbjct 291 TTESKLRREFEVYGPIKRIHMVYSKRSGKPRGYAFIEYEHERDMHSAYKHADGKKIDGRR 350
Query 92 VTVQM 96
V V +
Sbjct 351 VLVDV 355
> CE02197
Length=256
Score = 35.8 bits (81), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 14/50 (28%), Positives = 26/50 (52%), Gaps = 0/50 (0%)
Query 35 SDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNN 84
++R KIG+V +++ + P+ + F +E D RA+ ELN+
Sbjct 190 DELRDLFGKIGRVIRIFIARDKVTGLPKGFAFVTFESRDDAARAIAELND 239
> Hs14110420
Length=355
Score = 35.8 bits (81), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 21/86 (24%), Positives = 42/86 (48%), Gaps = 3/86 (3%)
Query 13 KTARTMRKFPYMLRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERE 72
KT ++K + G++P T +R + G+V+ + LP+ + R +CF ++ E
Sbjct 176 KTKEPVKKI--FVGGLSPDTPEEKIREYFGGFGEVESIELPMDNKTNKRRGFCFITFKEE 233
Query 73 SDGERAM-KELNNQMLMGFRVTVQMA 97
++ M K+ +N L + V M+
Sbjct 234 EPVKKIMEKKYHNVGLSKCEIKVAMS 259
> Hs22050771
Length=437
Score = 35.8 bits (81), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 30/65 (46%), Gaps = 0/65 (0%)
Query 32 TTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGFR 91
TT S +R G +K +++ PR Y F +YE E D A K + + + G R
Sbjct 114 TTESKLRREFEVYGPIKRIHMVYSKRSGKPRGYAFIEYEHERDMHSAYKHADGKKIDGRR 173
Query 92 VTVQM 96
V V +
Sbjct 174 VLVDV 178
> YIR001c
Length=250
Score = 35.8 bits (81), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 32/68 (47%), Gaps = 1/68 (1%)
Query 27 GVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQM 86
+ P TP + GQ+K + L TP+ Y + ++E + E+A+ +LN
Sbjct 70 NITPDVTPEQIEDHFKDCGQIKRITLLYDRNTGTPKGYGYIEFESPAYREKAL-QLNGGE 128
Query 87 LMGFRVTV 94
L G ++ V
Sbjct 129 LKGKKIAV 136
> Hs14110417
Length=336
Score = 35.8 bits (81), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 21/86 (24%), Positives = 42/86 (48%), Gaps = 3/86 (3%)
Query 13 KTARTMRKFPYMLRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERE 72
KT ++K + G++P T +R + G+V+ + LP+ + R +CF ++ E
Sbjct 157 KTKEPVKKI--FVGGLSPDTPEEKIREYFGGFGEVESIELPMDNKTNKRRGFCFITFKEE 214
Query 73 SDGERAM-KELNNQMLMGFRVTVQMA 97
++ M K+ +N L + V M+
Sbjct 215 EPVKKIMEKKYHNVGLSKCEIKVAMS 240
> At5g37720
Length=292
Score = 35.8 bits (81), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 37/81 (45%), Gaps = 3/81 (3%)
Query 31 GTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMG- 89
G T D+R S+IG+V E Y + P Y R SD +A+K+ NN +L G
Sbjct 107 GVTNEDIRELFSEIGEV-ERYAIHYDKNGRPSGTAEVVYPRRSDAFQALKKYNNVLLDGR 165
Query 90 -FRVTVQMANSDPKTPEQMRT 109
R+ + N+ + P R
Sbjct 166 PMRLEILGGNNSSEAPLSGRV 186
> Hs5454064
Length=669
Score = 35.8 bits (81), Expect = 0.021, Method: Composition-based stats.
Identities = 24/101 (23%), Positives = 47/101 (46%), Gaps = 9/101 (8%)
Query 2 RAGELWLVENAKTARTMRKFPYMLRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTP 61
R G +VE ++ R + + + V+ T ++RS + G+V E
Sbjct 61 RPGRALVVEMSR-PRPLNTWKIFVGNVSAACTSQELRSLFERRGRVIEC--------DVV 111
Query 62 RSYCFFDYERESDGERAMKELNNQMLMGFRVTVQMANSDPK 102
+ Y F E+E+D + A+ +LN + + G R+ V+++ K
Sbjct 112 KDYAFVHMEKEADAKAAIAQLNGKEVKGKRINVELSTKGQK 152
> At5g41690
Length=599
Score = 35.4 bits (80), Expect = 0.022, Method: Composition-based stats.
Identities = 21/88 (23%), Positives = 44/88 (50%), Gaps = 1/88 (1%)
Query 28 VAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQML 87
++P T SD+ F + +G+V + L V+H E Y F ++ + ++A++ N + L
Sbjct 179 LSPQTKISDIFDFFNCVGEVVSIRLMVNH-EGKHVGYGFVEFASADETKKALENKNGEYL 237
Query 88 MGFRVTVQMANSDPKTPEQMRTIYERKD 115
++ + +A + P P Y R++
Sbjct 238 HDHKIFIDVAKTAPYPPGPNYEDYLRRE 265
> CE00983
Length=456
Score = 35.4 bits (80), Expect = 0.024, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 29/67 (43%), Gaps = 0/67 (0%)
Query 31 GTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDYERESDGERAMKELNNQMLMGF 90
G T +VRS + IG+++ L + Y F +Y RE D RA+ N L
Sbjct 52 GMTQEEVRSLFTSIGEIESCKLVRDKVTGQSLGYGFVNYVREEDALRAVSSFNGLRLQNK 111
Query 91 RVTVQMA 97
+ V A
Sbjct 112 TIKVSYA 118
> CE21988
Length=434
Score = 35.4 bits (80), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 21/78 (26%), Positives = 31/78 (39%), Gaps = 0/78 (0%)
Query 10 ENAKTARTMRKFPYMLRGVAPGTTPSDVRSFLSKIGQVKEMYLPVHHLERTPRSYCFFDY 69
EN T R F + + + +R K G+V E + + + Y F Y
Sbjct 121 ENRSKPETSRHFHVFVGDLCSEIDSTKLREAFVKFGEVSEAKIIRDNNTNKGKGYGFVSY 180
Query 70 ERESDGERAMKELNNQML 87
R D ERA+ E+N L
Sbjct 181 PRREDAERAIDEMNGAWL 198
Lambda K H
0.319 0.132 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167969826
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40