bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
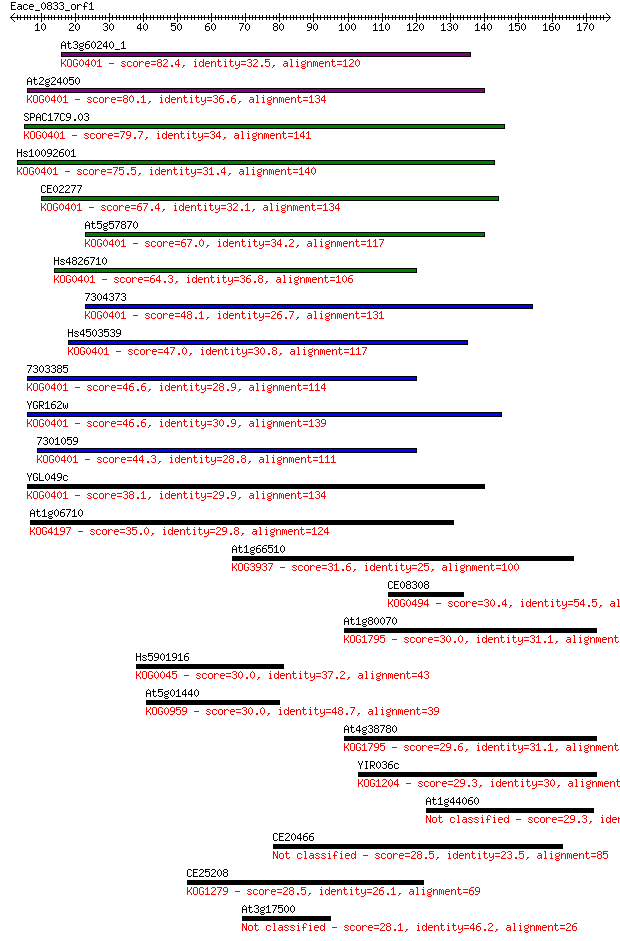
Query= Eace_0833_orf1
Length=176
Score E
Sequences producing significant alignments: (Bits) Value
At3g60240_1 82.4 4e-16
At2g24050 80.1 2e-15
SPAC17C9.03 79.7 3e-15
Hs10092601 75.5 5e-14
CE02277 67.4 2e-11
At5g57870 67.0 2e-11
Hs4826710 64.3 1e-10
7304373 48.1 9e-06
Hs4503539 47.0 2e-05
7303385 46.6 3e-05
YGR162w 46.6 3e-05
7301059 44.3 1e-04
YGL049c 38.1 0.009
At1g06710 35.0 0.078
At1g66510 31.6 0.93
CE08308 30.4 2.1
At1g80070 30.0 2.2
Hs5901916 30.0 2.4
At5g01440 30.0 2.6
At4g38780 29.6 3.1
YIR036c 29.3 4.4
At1g44060 29.3 4.6
CE20466 28.5 6.4
CE25208 28.5 8.0
At3g17500 28.1 9.8
> At3g60240_1
Length=1528
Score = 82.4 bits (202), Expect = 4e-16, Method: Composition-based stats.
Identities = 39/121 (32%), Positives = 78/121 (64%), Gaps = 2/121 (1%)
Query 16 RIKRRVLGVVKLIGELFHRKLLGFKIVNDVVVELVMRSEEPDEYLVECFLQLISTVGFFI 75
+++RR+LG ++LIGEL+ +++L KI++ + +L+ +++P E +E +L+ST+G I
Sbjct 1034 QVRRRMLGNIRLIGELYKKRMLTEKIMHACIQKLLGYNQDPHEENIEALCKLMSTIGVMI 1093
Query 76 DQNPKMKVVLDSWFGRLKELQTKN-YSKRLKCVIQDTFDMRRADWRKKIHRERAKALNEL 134
D N K K +D +F ++K L K S R++ ++ + D+R+ W++++ E K + E+
Sbjct 1094 DHN-KAKFQMDGYFEKMKMLSCKQELSSRVRFMLINAIDLRKNKWQERMKVEGPKKIEEV 1152
Query 135 H 135
H
Sbjct 1153 H 1153
> At2g24050
Length=747
Score = 80.1 bits (196), Expect = 2e-15, Method: Composition-based stats.
Identities = 49/137 (35%), Positives = 78/137 (56%), Gaps = 4/137 (2%)
Query 6 ESEEMQQEATRIKRRVLGVVKLIGELFHRKLLGFKIVNDVVVELVMRSEE--PDEYLVEC 63
E E M +E K R LG ++LIGEL +K++ KIV+ +V EL+ + P E VE
Sbjct 286 EMERMDKEKM-AKLRTLGNIRLIGELLKQKMVPEKIVHHIVQELLGDDTKACPAEGDVEA 344
Query 64 FLQLISTVGFFIDQNPKMKVVLDSWFGRLKEL-QTKNYSKRLKCVIQDTFDMRRADWRKK 122
Q T+G +D +P+ + + D++FGRLKEL + RL+ ++Q+ D+R W +
Sbjct 345 LCQFFITIGKQLDDSPRSRGINDTYFGRLKELARHPQLELRLRFMVQNVVDLRANKWVPR 404
Query 123 IHRERAKALNELHDQLE 139
+AK +NE+H + E
Sbjct 405 REEVKAKKINEIHSEAE 421
> SPAC17C9.03
Length=1403
Score = 79.7 bits (195), Expect = 3e-15, Method: Composition-based stats.
Identities = 48/146 (32%), Positives = 79/146 (54%), Gaps = 5/146 (3%)
Query 5 GESEEMQQE---ATRIKRRVLGVVKLIGELFHRKLLGFKIVNDVVVELVMRSEEPDEYLV 61
GE+E M E A IKRR LG+V+ IGELF +L KI+++ + L+ +P+E +
Sbjct 1128 GEAEIMSDEYYVAAAIKRRGLGLVRFIGELFKLSMLSEKIMHECIKRLLGNVTDPEEEEI 1187
Query 62 ECFLQLISTVGFFIDQNPKMKVVLDSWFGRLKEL-QTKNYSKRLKCVIQDTFDMRRADWR 120
E +L+ TVG ID K +D + R++ + + N R+K ++ D D R+ W
Sbjct 1188 ESLCRLLMTVGVNIDATEKGHAAMDVYVLRMETITKIPNLPSRIKFMLMDVMDSRKNGWA 1247
Query 121 KKIHRERA-KALNELHDQLETEEVLG 145
K E+ K + E+H++ E ++ L
Sbjct 1248 VKNEVEKGPKTIAEIHEEAERKKALA 1273
> Hs10092601
Length=1585
Score = 75.5 bits (184), Expect = 5e-14, Method: Composition-based stats.
Identities = 44/141 (31%), Positives = 84/141 (59%), Gaps = 8/141 (5%)
Query 3 VHGESEEMQQEATRIKRRVLGVVKLIGELFHRKLLGFKIVNDVVVELVMRSEEPDEYLVE 62
+H E EE + +A +RR +G +K IGELF K+L I++D VV+L+ + DE +E
Sbjct 874 LHDELEEAKDKA---RRRSIGNIKFIGELFKLKMLTEAIMHDCVVKLL---KNHDEESLE 927
Query 63 CFLQLISTVGFFIDQNPKMKVVLDSWFGRLKEL-QTKNYSKRLKCVIQDTFDMRRADWRK 121
C +L++T+G +D K K +D +F +++++ + + S R++ ++QD D+R +W
Sbjct 928 CLCRLLTTIGKDLDFE-KAKPRMDQYFNQMEKIVKERKTSSRIRFMLQDVIDLRLCNWVS 986
Query 122 KIHRERAKALNELHDQLETEE 142
+ + K + ++H + + EE
Sbjct 987 RRADQGPKTIEQIHKEAKIEE 1007
> CE02277
Length=1156
Score = 67.4 bits (163), Expect = 2e-11, Method: Composition-based stats.
Identities = 43/144 (29%), Positives = 75/144 (52%), Gaps = 12/144 (8%)
Query 10 MQQEATRIKRRVLGVVKLIGELFHRKLLGFKIVNDVVVELV-------MRSEEPDEYLVE 62
+ +E + +RR GV+ +G L+ +LL KIV EL ++ E+ DE +
Sbjct 633 LAEEKQKFRRRKFGVMTFMGYLYRNQLLSTKIVQTCTFELFNSIKDQDIKKEDVDEESIH 692
Query 63 CFLQLISTVGFFIDQNP-KMKVVLDSWFGRLKELQTKNY-SKRLKCVIQDTFDMRRADWR 120
C LQLI TVG +D++ V LD WF +L+ K Y S +++ +I + ++R+ W
Sbjct 693 CGLQLIETVGVMLDKSKDSTTVFLDQWFQKLE--AAKPYCSNKIRFMIMNLMELRKDKWI 750
Query 121 KKIHRERA-KALNELHDQLETEEV 143
+ E K ++E+H + E++
Sbjct 751 PRKSTESGPKKIDEIHKDIRQEKI 774
> At5g57870
Length=780
Score = 67.0 bits (162), Expect = 2e-11, Method: Composition-based stats.
Identities = 40/120 (33%), Positives = 65/120 (54%), Gaps = 3/120 (2%)
Query 23 GVVKLIGELFHRKLLGFKIVNDVVVELVMRSEE--PDEYLVECFLQLISTVGFFIDQNPK 80
G ++LIGEL +K++ KIV+ +V EL+ E+ P E VE T+G +D N K
Sbjct 343 GNIRLIGELLKQKMVPEKIVHHIVQELLGADEKVCPAEENVEAICHFFKTIGKQLDGNVK 402
Query 81 MKVVLDSWFGRLKEL-QTKNYSKRLKCVIQDTFDMRRADWRKKIHRERAKALNELHDQLE 139
K + D +F RL+ L + RL+ ++Q+ DMR W + +A+ + E+H + E
Sbjct 403 SKRINDVYFKRLQALSKNPQLELRLRFMVQNIIDMRSNGWVPRREEMKARTITEIHTEAE 462
> Hs4826710
Length=1396
Score = 64.3 bits (155), Expect = 1e-10, Method: Composition-based stats.
Identities = 39/108 (36%), Positives = 67/108 (62%), Gaps = 6/108 (5%)
Query 14 ATRIKRRV-LGVVKLIGELFHRKLLGFKIVNDVVVELVMRSEEPDEYLVECFLQLISTVG 72
A I RR LG +K IGELF K+L I++D VV+L+ + DE +EC +L++T+G
Sbjct 691 ARDIARRCSLGNIKFIGELFKLKMLTEAIMHDCVVKLL---KNHDEESLECLCRLLTTIG 747
Query 73 FFIDQNPKMKVVLDSWFGRLKE-LQTKNYSKRLKCVIQDTFDMRRADW 119
+D K K +D +F ++++ ++ K S R++ ++QD D+R ++W
Sbjct 748 KDLDFE-KAKPRMDQYFNQMEKIIKEKKTSSRIRFMLQDVLDLRGSNW 794
> 7304373
Length=1666
Score = 48.1 bits (113), Expect = 9e-06, Method: Composition-based stats.
Identities = 35/143 (24%), Positives = 68/143 (47%), Gaps = 22/143 (15%)
Query 23 GVVKLIGELFHRKLLGFKIVNDVVVELVMRSEEPDEYLVECFLQLISTVGFFID------ 76
G V+ IGELF +L KI+ + L+ E ++EC +L++TVG +
Sbjct 895 GTVRFIGELFKISMLTGKIIYSCIDTLLNPHSED---MLECLCKLLTTVGAKFEKTPVNS 951
Query 77 QNPKMKVVLDSWFGRLKELQTKN------YSKRLKCVIQDTFDMRRADWRKKIHRERAKA 130
++P L+ R++ + +K S R++ ++QD D+R+ W+ E K
Sbjct 952 KDPSRCYSLEKSITRMQAIASKTDKDGARVSSRVRFMLQDVIDLRKNKWQTS-RNEAPKT 1010
Query 131 LNELHDQLETEEVLGGAVHAAQY 153
+ ++ + + E++ +AQY
Sbjct 1011 MGQIEKEAKNEQL------SAQY 1027
> Hs4503539
Length=907
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 36/125 (28%), Positives = 65/125 (52%), Gaps = 10/125 (8%)
Query 18 KRRVLGVVKLIGELFHRKLLGFKIVNDVVVELVMRS-----EEPDEYLVECFLQLISTVG 72
K ++LG +K IGEL L+ I++ + L+ + ++ E L EC Q++ TVG
Sbjct 204 KIKMLGNIKFIGELGKLDLIHESILHKCIKTLLEKKKRVQLKDMGEDL-ECLCQIMRTVG 262
Query 73 FFIDQNPKMKVVLDSWFGRLKELQ-TKNYSKRLKCVIQDTFDMRRADW--RKKIHRERAK 129
+D + K ++D +F R+ L +K R++ ++QDT ++R W RK K
Sbjct 263 PRLDHE-RAKSLMDQYFARMCSLMLSKELPARIRFLLQDTVELREHHWVPRKAFLDNGPK 321
Query 130 ALNEL 134
+N++
Sbjct 322 TINQI 326
> 7303385
Length=869
Score = 46.6 bits (109), Expect = 3e-05, Method: Composition-based stats.
Identities = 33/122 (27%), Positives = 67/122 (54%), Gaps = 9/122 (7%)
Query 6 ESEEMQQEATRI-KRRVLGVVKLIGELFHRKLLGFKIVNDVVVELV----MRSEEPDEYL 60
E+E ++E + K+R+LG VK IGEL +L +++ ++EL R+ E
Sbjct 54 ENEADEEERRHLAKQRMLGNVKFIGELNKLDMLSKNVLHQCIMELFDKKKKRTAGTQEMC 113
Query 61 --VECFLQLISTVGFFIDQNPKMKVVLDSWFGRL-KELQTKNYSKRLKCVIQDTFDMRRA 117
+EC QL+ T G +D + K +++ +F +L + ++ Y R++ +++D ++R+
Sbjct 114 EDMECLAQLLKTCGKNLDSE-QGKELMNQYFEKLERRSKSSEYPPRIRFMLKDVIELRQN 172
Query 118 DW 119
+W
Sbjct 173 NW 174
> YGR162w
Length=952
Score = 46.6 bits (109), Expect = 3e-05, Method: Composition-based stats.
Identities = 43/153 (28%), Positives = 74/153 (48%), Gaps = 14/153 (9%)
Query 6 ESEEMQQE---ATRIKRRVLGVVKLIGELFHRKLLGFKIVNDVVVELVMR-SEEPDEYLV 61
E E M +E A KRR LG+V+ IG L+ LL K++ + L+ ++ P E +
Sbjct 725 EPEMMSEEYYAAASAKRRGLGLVRFIGFLYRLNLLTGKMMFECFRRLMKDLTDSPSEETL 784
Query 62 ECFLQLISTVGFFIDQNP--------KMKVVLDSWFGRLKE-LQTKNYSKRLKCVIQDTF 112
E ++L++TVG + + + +LDS FG L +QT S R+K + D
Sbjct 785 ESVVELLNTVGEQFETDSFRTGQATLEGSQLLDSLFGILDNIIQTAKISSRIKFKLIDIK 844
Query 113 DMRR-ADWRKKIHRERAKALNELHDQLETEEVL 144
++R +W K + ++H++ E + L
Sbjct 845 ELRHDKNWNSDKKDNGPKTIQQIHEEEERQRQL 877
> 7301059
Length=1905
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 32/128 (25%), Positives = 65/128 (50%), Gaps = 21/128 (16%)
Query 9 EMQQEATRIKRRVLGVVKLIGELFH-RKLLGFKIVNDVVVELVMRSEEPDEYLVECFLQL 67
EM+ + +RR G V+ IGELF + L +++N V L EE EY+ +L
Sbjct 1228 EMEDLEYQFRRRAWGTVRFIGELFKLQSLTNDRVLNCVESLLEHGCEEKLEYMC----KL 1283
Query 68 ISTVGFFID----QNPKMKVVLDSWFGRLKELQTKN------------YSKRLKCVIQDT 111
++TVG ++ ++ +++ ++ F R++++ ++ S R++ ++QD
Sbjct 1284 LTTVGHLLESSLPEHYQLRDRIEKIFRRIQDIINRSRGTSHRQQVHIKISSRVRFMMQDV 1343
Query 112 FDMRRADW 119
D+R +W
Sbjct 1344 LDLRLRNW 1351
> YGL049c
Length=914
Score = 38.1 bits (87), Expect = 0.009, Method: Composition-based stats.
Identities = 40/148 (27%), Positives = 69/148 (46%), Gaps = 14/148 (9%)
Query 6 ESEEMQQE---ATRIKRRVLGVVKLIGELFHRKLLGFKIVNDVVVELVMR-SEEPDEYLV 61
E E M E A KRR LG+V+ IG L+ LL K++ + L+ + +P E +
Sbjct 685 EPEMMSDEYYIAAAAKRRGLGLVRFIGYLYCLNLLTGKMMFECFRRLMKDLNNDPSEETL 744
Query 62 ECFLQLISTVG------FFIDQNPKMK--VVLDSWFGRLKE-LQTKNYSKRLKCVIQDTF 112
E ++L++TVG F+ ++ V+LD+ F L+ + S R+K + D
Sbjct 745 ESVIELLNTVGEQFEHDKFVTPQATLEGSVLLDNLFMLLQHIIDGGTISNRIKFKLIDVK 804
Query 113 DMRR-ADWRKKIHRERAKALNELHDQLE 139
++R W K + ++H + E
Sbjct 805 ELREIKHWNSAKKDAGPKTIQQIHQEEE 832
> At1g06710
Length=946
Score = 35.0 bits (79), Expect = 0.078, Method: Composition-based stats.
Identities = 37/139 (26%), Positives = 62/139 (44%), Gaps = 20/139 (14%)
Query 7 SEEMQQEATRIKRRVLGVVKLIGELFHRKLLGFK---IVNDVVVELVMRSEEPDEYLVEC 63
SE + E R+ R V+ ++ + +G+K V + +V+L++R + DE + E
Sbjct 90 SESLVIEVLRLIARPSAVISFF--VWAGRQIGYKHTAPVYNALVDLIVRDD--DEKVPEE 145
Query 64 FLQLI-----STVGFFID-------QNPKMKVVLDSWFGRLKELQTKNYSKRLKCVIQDT 111
FLQ I G F++ +N + L+ GRLK+ + + C+IQ
Sbjct 146 FLQQIRDDDKEVFGEFLNVLVRKHCRNGSFSIALEE-LGRLKDFRFRPSRSTYNCLIQAF 204
Query 112 FDMRRADWRKKIHRERAKA 130
R D IHRE + A
Sbjct 205 LKADRLDSASLIHREMSLA 223
> At1g66510
Length=399
Score = 31.6 bits (70), Expect = 0.93, Method: Compositional matrix adjust.
Identities = 25/106 (23%), Positives = 46/106 (43%), Gaps = 9/106 (8%)
Query 66 QLISTVGFFIDQNPKMKVV-----LDSWFGRLKELQTKNYSKRLKCVIQDTFDMRRADWR 120
+ T+GFF+D P +V D W ++ E + + YS+ ++ + FD +
Sbjct 64 EFSPTIGFFVDVAPSQVIVRKWNQQDEWLTKVSEEEEERYSQAVRSL---EFDKNLGPYN 120
Query 121 KKIHRERAKALNEL-HDQLETEEVLGGAVHAAQYGNIVVVGERTNL 165
K + E N + D +E E +GG + I+ G +T +
Sbjct 121 LKQYGEWRHLSNYITKDVVEKFEPVGGEITVTYESAILKGGPKTAM 166
> CE08308
Length=310
Score = 30.4 bits (67), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 12/22 (54%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 112 FDMRRADWRKKIHRERAKALNE 133
F RRA WR+K HR+R + NE
Sbjct 228 FQNRRAKWRRKEHRQRDRTRNE 249
> At1g80070
Length=2382
Score = 30.0 bits (66), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 35/75 (46%), Gaps = 1/75 (1%)
Query 99 NYSKRLKCVIQDTFDMRRADWRKKIHRERAKALNELHDQLETEEV-LGGAVHAAQYGNIV 157
+Y K LKC + + R +KK H R+ A + E + V +G V Y +
Sbjct 492 SYQKLLKCYVLNELHHRPPKAQKKKHLFRSLAATKFFQSTELDWVEVGLQVCRQGYNMLN 551
Query 158 VVGERTNLNGEYLDY 172
++ R NLN +LDY
Sbjct 552 LLIHRKNLNYLHLDY 566
> Hs5901916
Length=702
Score = 30.0 bits (66), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 25/48 (52%), Gaps = 5/48 (10%)
Query 38 GFKIVNDVVVELVMRSEEPD-----EYLVECFLQLISTVGFFIDQNPK 80
G K+ N V+ LV R + D + + CFL+L + FF+ +PK
Sbjct 636 GIKLNNKVMQVLVARYADDDLIIDFDSFISCFLRLKTMFTFFLTMDPK 683
> At5g01440
Length=307
Score = 30.0 bits (66), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 25/44 (56%), Gaps = 6/44 (13%)
Query 41 IVNDVVVELVMRSEEPDEYLVECFLQLISTVGFFI-----DQNP 79
I++D V +R EE YLVEC +LI VGF++ D NP
Sbjct 56 IISDSVYN-KLRIEEKLGYLVECETRLIHGVGFYVCVVSSDYNP 98
> At4g38780
Length=2352
Score = 29.6 bits (65), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 35/75 (46%), Gaps = 1/75 (1%)
Query 99 NYSKRLKCVIQDTFDMRRADWRKKIHRERAKALNELHDQLETEEV-LGGAVHAAQYGNIV 157
+Y K LKC + + R +KK H R+ A + E + V +G V Y +
Sbjct 464 SYQKLLKCYLLNELHHRPPKAQKKKHLFRSLAATKFFQSTELDWVEVGLQVCRQGYNMLN 523
Query 158 VVGERTNLNGEYLDY 172
++ R NLN +LDY
Sbjct 524 LLIHRKNLNYLHLDY 538
> YIR036c
Length=263
Score = 29.3 bits (64), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 33/73 (45%), Gaps = 3/73 (4%)
Query 103 RLKCVIQDTFDMR-RADWRKKIHRE--RAKALNELHDQLETEEVLGGAVHAAQYGNIVVV 159
R C+ D + + D R+ + + KAL +T +L V AA +V+
Sbjct 181 RAVCIAPGVVDTQMQKDIRETLGPQGMTPKALERFTQLYKTSSLLDPKVPAAVLAQLVLK 240
Query 160 GERTNLNGEYLDY 172
G +LNG+YL Y
Sbjct 241 GIPDSLNGQYLRY 253
> At1g44060
Length=481
Score = 29.3 bits (64), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 25/49 (51%), Gaps = 6/49 (12%)
Query 123 IHRERAKALNELHDQLETEEVLGGAVHAAQYGNIVVVGERTNLNGEYLD 171
IH R K+ E+ D+L EE LG VH + V + T +G Y+D
Sbjct 263 IHYSRPKSYQEIQDEL--EEKLGRPVHLGE----VFIQTHTKSDGTYVD 305
> CE20466
Length=772
Score = 28.5 bits (62), Expect = 6.4, Method: Composition-based stats.
Identities = 20/92 (21%), Positives = 40/92 (43%), Gaps = 7/92 (7%)
Query 78 NPKMKVVLDSWFGRLKELQTKNYSKRLKCVIQDTFDMRRADWRKKIHRERA-------KA 130
+P + LD + +++ + T + L + Q F + DW KK+ +A K
Sbjct 268 DPSLVPKLDEYVSKIQSVGTSVNTSTLNALNQIVFADLKDDWVKKVVSRQADALLKAIKP 327
Query 131 LNELHDQLETEEVLGGAVHAAQYGNIVVVGER 162
L + ++ + V G+ + Y IV +G +
Sbjct 328 LRNVTEEFKKVSVAIGSASSDGYNEIVHIGTK 359
> CE25208
Length=789
Score = 28.5 bits (62), Expect = 8.0, Method: Composition-based stats.
Identities = 18/70 (25%), Positives = 36/70 (51%), Gaps = 6/70 (8%)
Query 53 SEEPDEYLVECFLQLISTVGFFIDQNPKMKVVLDSWFGRLKELQT-KNYSKRLKCVIQDT 111
S EP + ++ ++ IST G IDQ K + + R K ++++++ C++ +
Sbjct 214 SGEPPKEEIKTEIESISTPGLKIDQYQKQAIAM-----RTKGAPPGRDWTEQETCLLLEA 268
Query 112 FDMRRADWRK 121
+M + DW K
Sbjct 269 LEMFKDDWNK 278
> At3g17500
Length=438
Score = 28.1 bits (61), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 15/26 (57%), Gaps = 0/26 (0%)
Query 69 STVGFFIDQNPKMKVVLDSWFGRLKE 94
S V FFID+ KM V D + G + E
Sbjct 295 SCVSFFIDEEKKMAVFCDGYIGGINE 320
Lambda K H
0.321 0.138 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2707167450
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40