bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
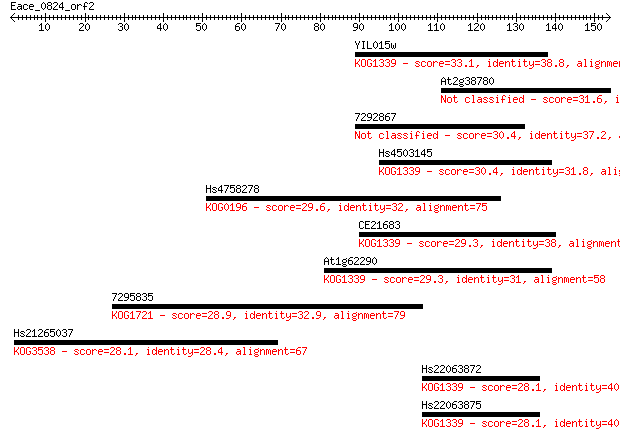
Query= Eace_0824_orf2
Length=153
Score E
Sequences producing significant alignments: (Bits) Value
YIL015w 33.1 0.19
At2g38780 31.6 0.54
7292867 30.4 1.3
Hs4503145 30.4 1.4
Hs4758278 29.6 2.5
CE21683 29.3 2.9
At1g62290 29.3 3.2
7295835 28.9 4.4
Hs21265037 28.1 6.5
Hs22063872 28.1 7.1
Hs22063875 28.1 7.4
> YIL015w
Length=587
Score = 33.1 bits (74), Expect = 0.19, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 28/52 (53%), Gaps = 7/52 (13%)
Query 89 PNLHFLVPESTHPFADLPFDGLVGLGFP---DVSGEEGLPSSALPIVDQMVK 137
PN+ F V + +A P G++G+GFP V G EG P+ P Q++K
Sbjct 153 PNIQFGVAK----YATTPVSGVLGIGFPRRESVKGYEGAPNEYYPNFPQILK 200
> At2g38780
Length=435
Score = 31.6 bits (70), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 27/47 (57%), Gaps = 5/47 (10%)
Query 111 VGLGFPDVSGEEGLPSSALPIVDQM----VKEVCYCCDETQSRFAQH 153
+GLGF D GLP+ +P+VD + + EV + + ++RFA+
Sbjct 149 MGLGFADKKSTRGLPAGLVPVVDYLPEGDLPEVEFIVGD-KTRFAEK 194
> 7292867
Length=153
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 24/43 (55%), Gaps = 8/43 (18%)
Query 89 PNLHFLVPESTHPFADLPFDGLVGLGFPDVSGEEGLPSSALPI 131
P LHF +P S+HP +PF LG P+ G+ L + +P+
Sbjct 62 PVLHFYIPSSSHP---IPF-----LGMPNTRGQTPLDAPCMPV 96
> Hs4503145
Length=396
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 14/44 (31%), Positives = 22/44 (50%), Gaps = 5/44 (11%)
Query 95 VPESTHPFADLPFDGLVGLGFPDVSGEEGLPSSALPIVDQMVKE 138
V E F D FDG++GLG+P ++ P+ D M+ +
Sbjct 169 VTEPGQTFVDAEFDGILGLGYPSLA-----VGGVTPVFDNMMAQ 207
> Hs4758278
Length=976
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 32/77 (41%), Gaps = 4/77 (5%)
Query 51 NHFVRSETPSFCSSAKCPTRQSAWQSKVRLQYFTTELKPNLHFLV--PESTHPFADLPFD 108
N R TP C SA WQ + + ++ L L+ P+S AD FD
Sbjct 831 NDGFRLPTPMDCPSAIYQLMMQCWQQERARRPKFADIVSILDKLIRAPDSLKTLAD--FD 888
Query 109 GLVGLGFPDVSGEEGLP 125
V + P SG EG+P
Sbjct 889 PRVSIRLPSTSGSEGVP 905
> CE21683
Length=320
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 30/53 (56%), Gaps = 3/53 (5%)
Query 90 NLHFLVPES-THPFADLPFDGLVGLGFPDVSGEEGLP--SSALPIVDQMVKEV 139
N F V E+ FA+ P DG++GLG+P ++ ++ P + LP +D V V
Sbjct 160 NQEFGVAETLASVFAEQPVDGILGLGWPALADDKVTPPMQNLLPQLDAKVFTV 212
> At1g62290
Length=526
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 30/58 (51%), Gaps = 14/58 (24%)
Query 81 QYFTTELKPNLHFLVPESTHPFADLPFDGLVGLGFPDVSGEEGLPSSALPIVDQMVKE 138
++ T +P L FLV + FDGL+GLGF +++ +A P+ M+K+
Sbjct 176 EFIETTSEPGLTFLVAK---------FDGLLGLGFQEIA-----VGNATPVWYNMLKQ 219
> 7295835
Length=345
Score = 28.9 bits (63), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 26/79 (32%), Positives = 30/79 (37%), Gaps = 13/79 (16%)
Query 27 FSTELEHAFFEWVATSWRSAACKKNHFVRSETPSFCSSAKCPTRQSAWQSKVRLQYFTTE 86
S+ EHA E + T R + H R +FCS A C VR Q T
Sbjct 175 MSSTSEHAVKEELRTKARQLSRNSTHTCRLCNKTFCSKASC----------VRHQKTHTG 224
Query 87 LKPNLHFLVPESTHPFADL 105
KP F PFADL
Sbjct 225 EKP---FACEICQKPFADL 240
> Hs21265037
Length=1205
Score = 28.1 bits (61), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 19/80 (23%), Positives = 32/80 (40%), Gaps = 15/80 (18%)
Query 2 ETQQSHSFKCVLHTSRLQEIPLNGLFSTELEHAFFEWVATSWRSA-------------AC 48
+T+ S ++K ++H + I N + EL+ FEW SW C
Sbjct 815 DTRSSLTYKYIIHEDSVPTINSNNVIQEELD--TFEWALKSWSQCSKPCGGGFQYTKYGC 872
Query 49 KKNHFVRSETPSFCSSAKCP 68
++ + SFC + K P
Sbjct 873 RRKSDNKMVHRSFCEANKKP 892
> Hs22063872
Length=325
Score = 28.1 bits (61), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 18/30 (60%), Gaps = 5/30 (16%)
Query 106 PFDGLVGLGFPDVSGEEGLPSSALPIVDQM 135
PFDG++GL +P +S S A P+ D +
Sbjct 115 PFDGILGLAYPSISS-----SGATPVFDNI 139
> Hs22063875
Length=325
Score = 28.1 bits (61), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 18/30 (60%), Gaps = 5/30 (16%)
Query 106 PFDGLVGLGFPDVSGEEGLPSSALPIVDQM 135
PFDG++GL +P +S S A P+ D +
Sbjct 115 PFDGILGLAYPSISS-----SGATPVFDNI 139
Lambda K H
0.321 0.132 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1961355924
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40