bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0777_orf2
Length=235
Score E
Sequences producing significant alignments: (Bits) Value
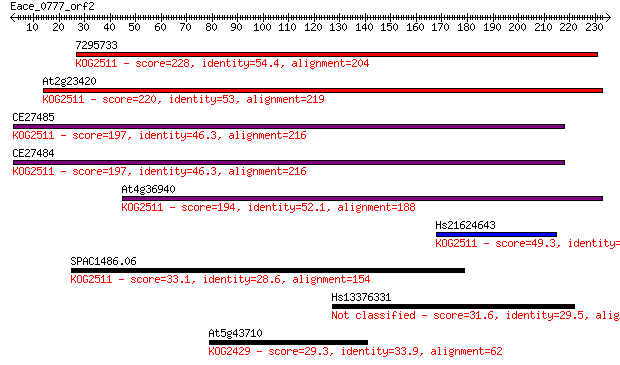
7295733 228 1e-59
At2g23420 220 2e-57
CE27485 197 2e-50
CE27484 197 2e-50
At4g36940 194 2e-49
Hs21624643 49.3 8e-06
SPAC1486.06 33.1 0.52
Hs13376331 31.6 1.5
At5g43710 29.3 6.6
> 7295733
Length=541
Score = 228 bits (580), Expect = 1e-59, Method: Compositional matrix adjust.
Identities = 111/205 (54%), Positives = 146/205 (71%), Gaps = 1/205 (0%)
Query 27 AKEGELAAFAAYAMTFPDNFLALVDTYNTLSSGIPNFLAVALAMFKLGAKPKGVRIDSGD 86
+ EGELAA +YA+ FPD F+ALVDTY+ SG+ NF AVALA+ LG G+RIDSGD
Sbjct 261 SSEGELAAMVSYAIAFPDGFMALVDTYDVKRSGLLNFSAVALALNDLGYHALGIRIDSGD 320
Query 87 LAYLSREARRMFKQCEEAFGFP-FGGLSIVLSNDLNEATITALNDEGHEADVFGIGTNVV 145
LAYLS AR F++ E F P F L+IV SND+NE TI +LN++GH+ D FGIGT++V
Sbjct 321 LAYLSCLARETFEKVAERFKVPWFNKLTIVASNDINEDTILSLNEQGHKIDCFGIGTHLV 380
Query 146 TCQAQPALGVVYKLVELEGKPCMKLSEDVEKTSLPTAKAAYRLYNKEGVPAVDLIQSASM 205
TCQ QPALG VYKLVE+ G+P +KLS+DVEK ++P K AYRLY+ +G +DL+Q S
Sbjct 381 TCQRQPALGCVYKLVEINGQPRIKLSQDVEKVTMPGNKNAYRLYSADGHALIDLLQKVSE 440
Query 206 PRPVCGEKLFCKDLYADKRRCFFSP 230
P P G+K+ C+ + + +R + P
Sbjct 441 PPPAVGQKVLCRHPFQESKRAYVIP 465
> At2g23420
Length=574
Score = 220 bits (560), Expect = 2e-57, Method: Compositional matrix adjust.
Identities = 116/237 (48%), Positives = 147/237 (62%), Gaps = 24/237 (10%)
Query 14 LFGVWPEANFHQMAKEGELAAFAAYAMTFPDNFLALVDTYNTLSSGIPNFLAVALAMFKL 73
L G++ E N + ELAAF +YA+ FP FLALVDTY+ + SGIPNF AVALA+
Sbjct 268 LSGIFSETN------QSELAAFTSYALAFPKTFLALVDTYDVMKSGIPNFCAVALALNDF 321
Query 74 GA-----------------KPKGVRIDSGDLAYLSREARRMFKQCEEAFGFP-FGGLSIV 115
G K G+R+DSGDLAYLSREAR F E P FG + +
Sbjct 322 GCDHMEFEMQLMFIVLRRYKALGIRLDSGDLAYLSREARNFFCTVERELKVPGFGKMVVT 381
Query 116 LSNDLNEATITALNDEGHEADVFGIGTNVVTCQAQPALGVVYKLVELEGKPCMKLSEDVE 175
SNDLNE TI ALN +GHE D FGIGT +VTC +Q ALG V+KLVE+ +P +KLSEDV
Sbjct 382 ASNDLNEETIDALNKQGHEVDAFGIGTYLVTCYSQAALGCVFKLVEINNQPRIKLSEDVT 441
Query 176 KTSLPTAKAAYRLYNKEGVPAVDLIQSASMPRPVCGEKLFCKDLYADKRRCFFSPRR 232
K S+P K +YRLY KEG P VD++ + P P GE+L C+ + + +R + P+R
Sbjct 442 KVSIPCKKRSYRLYGKEGYPLVDIMTGENEPPPKVGERLLCRHPFNESKRAYVVPQR 498
> CE27485
Length=531
Score = 197 bits (501), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 100/218 (45%), Positives = 136/218 (62%), Gaps = 2/218 (0%)
Query 2 DFPQAVMSARDLLFGVWPEANFHQMAKEGELAAFAAYAMTFPDNFLALVDTYNTLSSGIP 61
D Q M R L + +GEL+AF AYA+ FPD FLAL+DTY+ + SG+
Sbjct 255 DLFQISMEKRAWLLDQFSWKAALSEVSDGELSAFVAYAIAFPDTFLALIDTYDVIRSGVV 314
Query 62 NFLAVALAMFKLGAKPKGVRIDSGDLAYLSREARRMFKQCEEAFGFP--FGGLSIVLSND 119
NF+AV+LA+ LG + G RIDSGDL+YLS+E R F + G F +SIV SND
Sbjct 315 NFVAVSLALHDLGYRSMGCRIDSGDLSYLSKELRECFVKVSTLKGEYKFFEKMSIVASND 374
Query 120 LNEATITALNDEGHEADVFGIGTNVVTCQAQPALGVVYKLVELEGKPCMKLSEDVEKTSL 179
+NE TI +LND+ HE + FG+GT++VTCQ QPALG VYKLV +P +KLS+DV K ++
Sbjct 375 INEETIMSLNDQQHEINAFGVGTHLVTCQKQPALGCVYKLVAQSAQPKIKLSQDVTKITI 434
Query 180 PTAKAAYRLYNKEGVPAVDLIQSASMPRPVCGEKLFCK 217
P K YR++ K G +DL+ P P +++ C+
Sbjct 435 PGKKKCYRIFGKNGYAILDLMMLEDEPEPQPNQQILCR 472
> CE27484
Length=547
Score = 197 bits (500), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 100/218 (45%), Positives = 136/218 (62%), Gaps = 2/218 (0%)
Query 2 DFPQAVMSARDLLFGVWPEANFHQMAKEGELAAFAAYAMTFPDNFLALVDTYNTLSSGIP 61
D Q M R L + +GEL+AF AYA+ FPD FLAL+DTY+ + SG+
Sbjct 271 DLFQISMEKRAWLLDQFSWKAALSEVSDGELSAFVAYAIAFPDTFLALIDTYDVIRSGVV 330
Query 62 NFLAVALAMFKLGAKPKGVRIDSGDLAYLSREARRMFKQCEEAFGFP--FGGLSIVLSND 119
NF+AV+LA+ LG + G RIDSGDL+YLS+E R F + G F +SIV SND
Sbjct 331 NFVAVSLALHDLGYRSMGCRIDSGDLSYLSKELRECFVKVSTLKGEYKFFEKMSIVASND 390
Query 120 LNEATITALNDEGHEADVFGIGTNVVTCQAQPALGVVYKLVELEGKPCMKLSEDVEKTSL 179
+NE TI +LND+ HE + FG+GT++VTCQ QPALG VYKLV +P +KLS+DV K ++
Sbjct 391 INEETIMSLNDQQHEINAFGVGTHLVTCQKQPALGCVYKLVAQSAQPKIKLSQDVTKITI 450
Query 180 PTAKAAYRLYNKEGVPAVDLIQSASMPRPVCGEKLFCK 217
P K YR++ K G +DL+ P P +++ C+
Sbjct 451 PGKKKCYRIFGKNGYAILDLMMLEDEPEPQPNQQILCR 488
> At4g36940
Length=458
Score = 194 bits (492), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 98/193 (50%), Positives = 127/193 (65%), Gaps = 5/193 (2%)
Query 45 NFLALVDT----YNTLSSGIPNFLAVALAMFKLGAKPKGVRIDSGDLAYLSREARRMFKQ 100
+F+ LV T + SGIPNF AVALA+ +LG K G+R+DSGDLAYLS E R+ F
Sbjct 252 DFICLVQTCLTKIQVMKSGIPNFCAVALALNELGYKAVGIRLDSGDLAYLSTEVRKFFCA 311
Query 101 CEEAFGFP-FGGLSIVLSNDLNEATITALNDEGHEADVFGIGTNVVTCQAQPALGVVYKL 159
E P FG + + SNDLNE T+ ALN +GHE D FGIGTN+VTC AQ ALG V+KL
Sbjct 312 IERDLKVPDFGKMIVTASNDLNEETVDALNKQGHEVDAFGIGTNLVTCYAQAALGCVFKL 371
Query 160 VELEGKPCMKLSEDVEKTSLPTAKAAYRLYNKEGVPAVDLIQSASMPRPVCGEKLFCKDL 219
VE+ +P +KLSEDV K S+P K YRL+ KEG P VD++ + P P GE+L C+
Sbjct 372 VEINNQPRIKLSEDVTKVSIPCKKRTYRLFGKEGYPLVDIMTGENEPPPKVGERLLCRHP 431
Query 220 YADKRRCFFSPRR 232
+ + +R + P+R
Sbjct 432 FNESKRAYVVPQR 444
> Hs21624643
Length=133
Score = 49.3 bits (116), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 23/47 (48%), Positives = 33/47 (70%), Gaps = 0/47 (0%)
Query 168 MKLSEDVEKTSLPTAKAAYRLYNKEGVPAVDLIQSASMPRPVCGEKL 214
MKL+ED EK +LP +KAA+RL +G P +D++Q A P P G++L
Sbjct 1 MKLTEDPEKQTLPGSKAAFRLLGSDGSPLMDMLQLAEEPVPQAGQEL 47
> SPAC1486.06
Length=410
Score = 33.1 bits (74), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 44/169 (26%), Positives = 67/169 (39%), Gaps = 25/169 (14%)
Query 25 QMAKEGELAAFAAYAMTFPDNFL-ALVDTYNT---LSSGIPNFLAVALAMFKLGAKPKGV 80
Q K+ A + TF + L AL DT++T L S N +F GV
Sbjct 234 QNYKQANRIASLKWVQTFGTSLLIALTDTFSTDVFLKSFTANSADDLANVFH------GV 287
Query 81 RIDSGDLAYLSREARRMFKQCEEAFGFPFGGLSIVLSNDLNEATITALNDEGHEADV--- 137
R DSG + + +K + G IV S+ LN L + +
Sbjct 288 RQDSGCAEEYIEKVVKHYK----SIGVDPSTKVIVHSDALNVDRCIELYKYCEKCGIKSA 343
Query 138 FGIGTNVVT--------CQAQPALGVVYKLVELEGKPCMKLSEDVEKTS 178
FGIGTN+ + + + +V KL EG +K+S+D+ K +
Sbjct 344 FGIGTNLTSDFQKVSNPSEVSKPMNIVIKLFSAEGTKAVKISDDIMKNT 392
> Hs13376331
Length=463
Score = 31.6 bits (70), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 28/105 (26%), Positives = 46/105 (43%), Gaps = 16/105 (15%)
Query 127 ALNDEGHEADVFGIGT-------NVVTCQAQPALGVVYKLVELEG---KPCMKLSEDVEK 176
A++ GH + V G C AL +Y+ + +G K C KL VE+
Sbjct 86 AVHQTGHNSGVIHSGIYYKPESLKAKLCVQGAAL--LYEYCQQKGISYKQCGKLIVAVEQ 143
Query 177 TSLPTAKAAYRLYNKEGVPAVDLIQSASMPRPVCGEKLFCKDLYA 221
+P +A Y + GVP + LIQ + + ++ +C+ L A
Sbjct 144 EEIPRLQALYEKGLQNGVPGLRLIQQEDIKK----KEPYCRGLMA 184
> At5g43710
Length=624
Score = 29.3 bits (64), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 34/65 (52%), Gaps = 3/65 (4%)
Query 79 GVRIDS--GDLAYLSRE-ARRMFKQCEEAFGFPFGGLSIVLSNDLNEATITALNDEGHEA 135
G+RI S +L L+ ARRM + G PFG ++++ D +E+ IT+ G +
Sbjct 144 GMRIPSYNNELLVLAENLARRMLPAFDTPTGIPFGSVNLMYGVDKHESKITSTAGGGTLS 203
Query 136 DVFGI 140
FG+
Sbjct 204 LEFGV 208
Lambda K H
0.321 0.137 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4633705180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40