bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0759_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
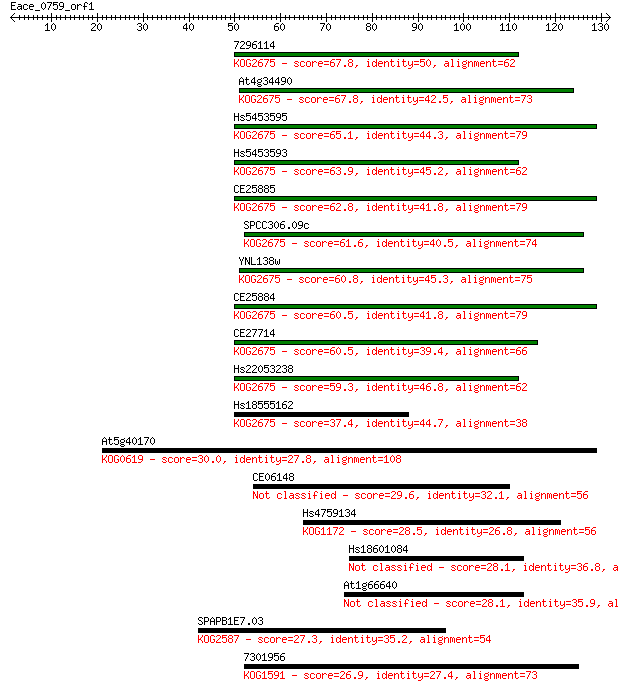
7296114 67.8 5e-12
At4g34490 67.8 5e-12
Hs5453595 65.1 3e-11
Hs5453593 63.9 8e-11
CE25885 62.8 2e-10
SPCC306.09c 61.6 3e-10
YNL138w 60.8 6e-10
CE25884 60.5 7e-10
CE27714 60.5 8e-10
Hs22053238 59.3 2e-09
Hs18555162 37.4 0.008
At5g40170 30.0 1.0
CE06148 29.6 1.5
Hs4759134 28.5 3.6
Hs18601084 28.1 3.9
At1g66640 28.1 4.9
SPAPB1E7.03 27.3 6.8
7301956 26.9 8.9
> 7296114
Length=521
Score = 67.8 bits (164), Expect = 5e-12, Method: Composition-based stats.
Identities = 31/62 (50%), Positives = 47/62 (75%), Gaps = 2/62 (3%)
Query 50 RLQVLGSAPSISIDKSQKVDLFLSNESREVEITSSKSSEMNLNVPLEGEEGDWQEIAIPE 109
++QVLGS P++SIDK+ ++LS +S VEI +SKSSEMN+ +P + GD+ E+A+PE
Sbjct 444 QMQVLGSVPTVSIDKTDGCQMYLSKDSLGVEIVNSKSSEMNILLP--DDSGDYTELALPE 501
Query 110 QF 111
Q+
Sbjct 502 QY 503
> At4g34490
Length=476
Score = 67.8 bits (164), Expect = 5e-12, Method: Composition-based stats.
Identities = 31/73 (42%), Positives = 49/73 (67%), Gaps = 1/73 (1%)
Query 51 LQVLGSAPSISIDKSQKVDLFLSNESREVEITSSKSSEMNLNVPLEGEEGDWQEIAIPEQ 110
+Q GSAP++S+D + L+L+ +S E IT++KSSE+N+ VP +GDW E A+P+Q
Sbjct 397 VQCQGSAPTVSVDNTTGCQLYLNKDSLETAITTAKSSEINVMVPGATPDGDWVEHALPQQ 456
Query 111 FHHKLKPDGKLHT 123
++H +GK T
Sbjct 457 YNHVFT-EGKFET 468
> Hs5453595
Length=475
Score = 65.1 bits (157), Expect = 3e-11, Method: Composition-based stats.
Identities = 35/79 (44%), Positives = 51/79 (64%), Gaps = 3/79 (3%)
Query 50 RLQVLGSAPSISIDKSQKVDLFLSNESREVEITSSKSSEMNLNVPLEGEEGDWQEIAIPE 109
++QV+G P+ISI+K+ +LS S + EI S+KSSEMN+ +P EG GD+ E +PE
Sbjct 398 KVQVMGKVPTISINKTDGCHAYLSKNSLDCEIVSAKSSEMNVLIPTEG--GDFNEFPVPE 455
Query 110 QFHHKLKPDGKLHTRVSDL 128
QF L KL T V+++
Sbjct 456 QF-KTLWNGQKLVTTVTEI 473
> Hs5453593
Length=477
Score = 63.9 bits (154), Expect = 8e-11, Method: Composition-based stats.
Identities = 28/62 (45%), Positives = 47/62 (75%), Gaps = 3/62 (4%)
Query 50 RLQVLGSAPSISIDKSQKVDLFLSNESREVEITSSKSSEMNLNVPLEGEEGDWQEIAIPE 109
++QV+G P+ISI+K++ ++LS ++ + EI S+KSSEMN+ +P ++GD++E IPE
Sbjct 401 QIQVMGRVPTISINKTEGCHIYLSEDALDCEIVSAKSSEMNILIP---QDGDYREFPIPE 457
Query 110 QF 111
QF
Sbjct 458 QF 459
> CE25885
Length=495
Score = 62.8 bits (151), Expect = 2e-10, Method: Composition-based stats.
Identities = 33/79 (41%), Positives = 51/79 (64%), Gaps = 2/79 (2%)
Query 50 RLQVLGSAPSISIDKSQKVDLFLSNESREVEITSSKSSEMNLNVPLEGEEGDWQEIAIPE 109
++Q LG P++SI K+ ++LS ++ EI +SKSSEMN++V ++GD+ E +PE
Sbjct 418 QIQTLGELPTLSIQKTDGCQVYLSKVAQGCEIVTSKSSEMNISVQ-TNDDGDYSEFPVPE 476
Query 110 QFHHKLKPDGKLHTRVSDL 128
QF +GKL T VSD+
Sbjct 477 QFKTTFV-NGKLVTVVSDI 494
> SPCC306.09c
Length=551
Score = 61.6 bits (148), Expect = 3e-10, Method: Composition-based stats.
Identities = 30/74 (40%), Positives = 45/74 (60%), Gaps = 2/74 (2%)
Query 52 QVLGSAPSISIDKSQKVDLFLSNESREVEITSSKSSEMNLNVPLEGEEGDWQEIAIPEQF 111
QV+ P I ID+ ++LS S E+ +SKS+ +N+NVP EEGD+ E A+PEQ
Sbjct 476 QVMNHVPMIVIDQCDGGSIYLSKSSLSSEVVTSKSTSLNINVP--NEEGDYAERAVPEQI 533
Query 112 HHKLKPDGKLHTRV 125
HK+ G+L + +
Sbjct 534 KHKVNEKGELVSEI 547
> YNL138w
Length=526
Score = 60.8 bits (146), Expect = 6e-10, Method: Composition-based stats.
Identities = 34/75 (45%), Positives = 47/75 (62%), Gaps = 2/75 (2%)
Query 51 LQVLGSAPSISIDKSQKVDLFLSNESREVEITSSKSSEMNLNVPLEGEEGDWQEIAIPEQ 110
+QV S P ISIDKS +++LS ES EI +S S+ +N+N+P+ GE+ D+ E IPEQ
Sbjct 449 IQVNHSLPQISIDKSDGGNIYLSKESLNTEIYTSCSTAINVNLPI-GEDDDYVEFPIPEQ 507
Query 111 FHHKLKPDGKLHTRV 125
H DGK + V
Sbjct 508 MKHSFA-DGKFKSAV 521
> CE25884
Length=1256
Score = 60.5 bits (145), Expect = 7e-10, Method: Composition-based stats.
Identities = 33/79 (41%), Positives = 51/79 (64%), Gaps = 2/79 (2%)
Query 50 RLQVLGSAPSISIDKSQKVDLFLSNESREVEITSSKSSEMNLNVPLEGEEGDWQEIAIPE 109
++Q LG P++SI K+ ++LS ++ EI +SKSSEMN++V ++GD+ E +PE
Sbjct 1179 QIQTLGELPTLSIQKTDGCQVYLSKVAQGCEIVTSKSSEMNISVQ-TNDDGDYSEFPVPE 1237
Query 110 QFHHKLKPDGKLHTRVSDL 128
QF +GKL T VSD+
Sbjct 1238 QFKTTFV-NGKLVTVVSDI 1255
> CE27714
Length=457
Score = 60.5 bits (145), Expect = 8e-10, Method: Composition-based stats.
Identities = 26/66 (39%), Positives = 45/66 (68%), Gaps = 0/66 (0%)
Query 50 RLQVLGSAPSISIDKSQKVDLFLSNESREVEITSSKSSEMNLNVPLEGEEGDWQEIAIPE 109
++Q LG P++SI K+ ++LS ++ +I +SKSSEMN++ LE + ++ E+A+PE
Sbjct 379 QIQTLGELPTVSIQKTDGCHIYLSRDALNAQIVASKSSEMNISAMLEDGDDEYTEMALPE 438
Query 110 QFHHKL 115
QF K+
Sbjct 439 QFMTKI 444
> Hs22053238
Length=264
Score = 59.3 bits (142), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 29/62 (46%), Positives = 42/62 (67%), Gaps = 2/62 (3%)
Query 50 RLQVLGSAPSISIDKSQKVDLFLSNESREVEITSSKSSEMNLNVPLEGEEGDWQEIAIPE 109
++QV+ P+ISI+K+ +LS S + EI S+KSSEMN+ +P EG GD+ E +PE
Sbjct 187 KVQVMSKVPTISINKTDGCHAYLSKNSLDCEIASAKSSEMNVLIPTEG--GDFNEFPVPE 244
Query 110 QF 111
QF
Sbjct 245 QF 246
> Hs18555162
Length=158
Score = 37.4 bits (85), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 26/38 (68%), Gaps = 0/38 (0%)
Query 50 RLQVLGSAPSISIDKSQKVDLFLSNESREVEITSSKSS 87
++QV+G P+ISI+K+ +LS S + EI S+KSS
Sbjct 121 KVQVMGKVPTISINKTDGCHAYLSKNSLDCEIVSAKSS 158
> At5g40170
Length=792
Score = 30.0 bits (66), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 52/114 (45%), Gaps = 6/114 (5%)
Query 21 FLLSSFFCLT-SSASSFIAVRFAFP-PYIFRRLQVLGSAPSISIDKSQKVDLFLSNESRE 78
F ++ FFC ++ S + F FP + L++ PS + D + K+D F ++R
Sbjct 8 FFITCFFCCVFVTSDSVYTLPFPFPRDQVEILLELKNEFPSFNCDLTWKLDYFGRMDTRA 67
Query 79 VEITSSKSSEMNLNVPLEGEEGDWQEIAIPEQFHHKLKPDGKL----HTRVSDL 128
+ +K S+ V + E G +E+++ Q LK + L H R DL
Sbjct 68 NISSWTKDSDSFSGVSFDSETGVVKELSLGRQCLTSLKANSSLFRFQHLRYLDL 121
> CE06148
Length=139
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 32/56 (57%), Gaps = 8/56 (14%)
Query 54 LGSAPSISIDKSQKVDLFLSNESREVEITSSKSSEMNLNVPLEGEEGDWQEIAIPE 109
L +AP+ + KS D S++ ++ SSK E P + +EG+++E+AIP+
Sbjct 92 LMAAPAGGVGKSDSFD------SKKQDVPSSKKDEPV--APKKEDEGNYEELAIPQ 139
> Hs4759134
Length=1044
Score = 28.5 bits (62), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 15/56 (26%), Positives = 27/56 (48%), Gaps = 0/56 (0%)
Query 65 SQKVDLFLSNESREVEITSSKSSEMNLNVPLEGEEGDWQEIAIPEQFHHKLKPDGK 120
SQ+V L E E + +E++ EGE+ +W+E A +F ++ G+
Sbjct 96 SQRVQFILGTEEDEEHVPHELFTELDEICMKEGEDAEWKETARWLKFEEDVEDGGE 151
> Hs18601084
Length=608
Score = 28.1 bits (61), Expect = 3.9, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Query 75 ESREVEITSSKSSEMNLNVPLEGEEGDWQEIAIPEQFH 112
++ ++ I +KS + +N LE +EGD I IP +FH
Sbjct 341 DTFDIRILMAKSVKYTVNF-LEAKEGDLHRIEIPFKFH 377
> At1g66640
Length=416
Score = 28.1 bits (61), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 21/39 (53%), Gaps = 2/39 (5%)
Query 74 NESREVEITSSKSSEMNLNVPLEGEEGDWQEIAIPEQFH 112
+E E + + +SS L+ G+EGDW+ IPE H
Sbjct 2 DEDGEKHVLTKRSSSPELSDKRSGDEGDWRR--IPEWRH 38
> SPAPB1E7.03
Length=591
Score = 27.3 bits (59), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 30/58 (51%), Gaps = 4/58 (6%)
Query 42 AFPPYIFRRLQVLGSAPSISIDKSQKVD----LFLSNESREVEITSSKSSEMNLNVPL 95
+ P RR + +GSAPS+S D + +D + + + S V I S K S ++ N L
Sbjct 224 SIPHPTKRRKRAVGSAPSVSTDLNNILDDDNSILVPDLSAHVRINSGKLSVLSKNARL 281
> 7301956
Length=495
Score = 26.9 bits (58), Expect = 8.9, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 35/74 (47%), Gaps = 1/74 (1%)
Query 52 QVLGSAPSISIDKSQKVDLFLSNESREVEITSSKSSEMNLNVPLEGEE-GDWQEIAIPEQ 110
Q L S S + ++ + FL+ S + + SK + +P EGE G++Q ++
Sbjct 204 QELIGIQSASEEHAKNYETFLNALSEKALLNESKPILEHAPIPEEGEPVGEFQAYSLTCS 263
Query 111 FHHKLKPDGKLHTR 124
H +L P + H R
Sbjct 264 GHWRLTPKEQRHLR 277
Lambda K H
0.324 0.139 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1283105810
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40