bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0748_orf1
Length=99
Score E
Sequences producing significant alignments: (Bits) Value
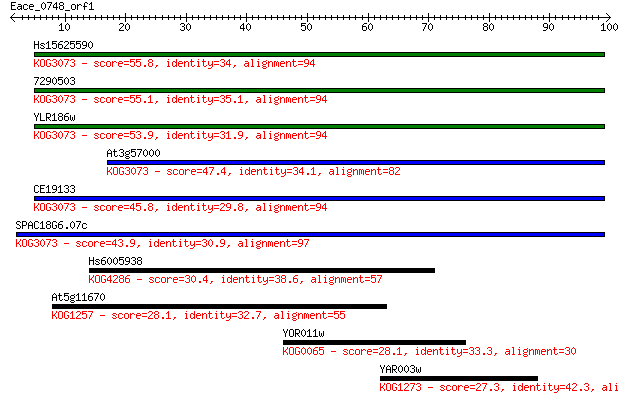
Hs15625590 55.8 2e-08
7290503 55.1 3e-08
YLR186w 53.9 7e-08
At3g57000 47.4 6e-06
CE19133 45.8 2e-05
SPAC18G6.07c 43.9 8e-05
Hs6005938 30.4 0.80
At5g11670 28.1 4.0
YOR011w 28.1 4.2
YAR003w 27.3 7.5
> Hs15625590
Length=151
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 32/94 (34%), Positives = 53/94 (56%), Gaps = 5/94 (5%)
Query 5 LLQVTKNDPNLFLPIGSRKIAFSSKGRHVDLENFVQQFQHTENPVLFLVGAVAHANPTAN 64
LL+V KN + P+G K+ S + + + V++ + +P++F+VGA AH +
Sbjct 62 LLKVIKNPVSDHFPVGCMKVGTSFS---IPVVSDVRELVPSSDPIVFVVGAFAHGK--VS 116
Query 65 NELAEECISISPCGLSAAVCCSSLCTEFENLWNI 98
E E+ +SIS LSAA+ C+ L T FE +W +
Sbjct 117 VEYTEKMVSISNYPLSAALTCAKLTTAFEEVWGV 150
> 7290503
Length=233
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 33/99 (33%), Positives = 51/99 (51%), Gaps = 7/99 (7%)
Query 5 LLQVTKNDPNLFLPIGSRKIAFSSKGRHV-DLENFVQQFQHT----ENPVLFLVGAVAHA 59
L+ V KN +P+G +K A S G+ + + + V T + PV+ ++GA AH
Sbjct 136 LMSVIKNPITDHVPVGCKKYAMSFSGKLLPNCRDLVPHGDETSASYDEPVVIVIGAFAHG 195
Query 60 NPTANNELAEECISISPCGLSAAVCCSSLCTEFENLWNI 98
+ EE SIS LSAA+ CS +C+ FE +W +
Sbjct 196 --VLKTDYTEELFSISNYPLSAAIACSKICSAFEEVWGV 232
> YLR186w
Length=252
Score = 53.9 bits (128), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 30/94 (31%), Positives = 49/94 (52%), Gaps = 1/94 (1%)
Query 5 LLQVTKNDPNLFLPIGSRKIAFSSKGRHVDLENFVQQFQHTENPVLFLVGAVAHANPTAN 64
LL+V KN LP RK+ S + +++++++ E+ +F VGA+A
Sbjct 159 LLKVIKNPITDHLPTKCRKVTLSFDAPVIRVQDYIEKLDDDESICVF-VGAMARGKDNFA 217
Query 65 NELAEECISISPCGLSAAVCCSSLCTEFENLWNI 98
+E +E + +S LSA+V CS C E+ WNI
Sbjct 218 DEYVDEKVGLSNYPLSASVACSKFCHGAEDAWNI 251
> At3g57000
Length=298
Score = 47.4 bits (111), Expect = 6e-06, Method: Composition-based stats.
Identities = 28/83 (33%), Positives = 43/83 (51%), Gaps = 3/83 (3%)
Query 17 LPIGSRKIAFS-SKGRHVDLENFVQQFQHTENPVLFLVGAVAHANPTANNELAEECISIS 75
LP+ S +I FS S + V+++ + + +F+VGA+AH N +E +S+S
Sbjct 217 LPVNSHRIGFSHSSEKLVNMQKHLATVCDDDRDTVFVVGAMAHGKIDCN--YIDEFVSVS 274
Query 76 PCGLSAAVCCSSLCTEFENLWNI 98
LSAA C S +C WNI
Sbjct 275 EYPLSAAYCISRICEALATNWNI 297
> CE19133
Length=231
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 48/94 (51%), Gaps = 3/94 (3%)
Query 5 LLQVTKNDPNLFLPIGSRKIAFSSKGRHVDLENFVQQFQHTENPVLFLVGAVAHANPTAN 64
L+ V KN + LP+GSRK+ S + + N + T+ P++ ++G +A +
Sbjct 140 LMSVVKNPVSNHLPVGSRKMLMSFNVPELTMANKLVA-PETDEPLVLIIGGIARGKIVVD 198
Query 65 NELAEECISISPCGLSAAVCCSSLCTEFENLWNI 98
+E IS P LSAA+ C+ + + E +W I
Sbjct 199 YNDSETKISNYP--LSAALTCAKVTSGLEEIWGI 230
> SPAC18G6.07c
Length=359
Score = 43.9 bits (102), Expect = 8e-05, Method: Composition-based stats.
Identities = 30/97 (30%), Positives = 45/97 (46%), Gaps = 1/97 (1%)
Query 2 NVTLLQVTKNDPNLFLPIGSRKIAFSSKGRHVDLENFVQQFQHTENPVLFLVGAVAHANP 61
N LL+V KN +LP RK S V +++ Q ++ V +GA+AH
Sbjct 263 NEKLLKVIKNPVTDYLPPNCRKATLSFDAPTVPPRKYLETLQPNQS-VCIAIGAMAHGPD 321
Query 62 TANNELAEECISISPCGLSAAVCCSSLCTEFENLWNI 98
++ +E ISIS LSA++ CS E+ I
Sbjct 322 DFSDGWVDEKISISDYPLSASIACSKFLHSMEDFLGI 358
> Hs6005938
Length=3433
Score = 30.4 bits (67), Expect = 0.80, Method: Composition-based stats.
Identities = 22/62 (35%), Positives = 29/62 (46%), Gaps = 5/62 (8%)
Query 14 NLFLPIGSRKIAFSSKGRHV-----DLENFVQQFQHTENPVLFLVGAVAHANPTANNELA 68
NL I R+ A ++ R V DLENF++ Q E V LV A N ++ LA
Sbjct 2426 NLKQSIADRQNALEAEWRTVQASRRDLENFLKWIQEAETTVNVLVDASHRENALQDSILA 2485
Query 69 EE 70
E
Sbjct 2486 RE 2487
> At5g11670
Length=588
Score = 28.1 bits (61), Expect = 4.0, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 29/55 (52%), Gaps = 3/55 (5%)
Query 8 VTKNDPNLFLPIGSRKIAFSSKGRHVDLENFVQQFQHTENPVLFLVGAVAHANPT 62
+T+ ++L + S+ + SS R L++F Q + H PV L+GAV PT
Sbjct 359 ITETRKKIWL-VDSKGLIVSS--RKESLQHFKQPWAHEHKPVKDLIGAVNAIKPT 410
> YOR011w
Length=1394
Score = 28.1 bits (61), Expect = 4.2, Method: Composition-based stats.
Identities = 10/30 (33%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 46 ENPVLFLVGAVAHANPTANNELAEECISIS 75
+NP F++ AV ++N +A + AEE ++++
Sbjct 984 DNPADFVIDAVGNSNSSAGKDTAEEALTLN 1013
> YAR003w
Length=426
Score = 27.3 bits (59), Expect = 7.5, Method: Composition-based stats.
Identities = 11/26 (42%), Positives = 15/26 (57%), Gaps = 0/26 (0%)
Query 62 TANNELAEECISISPCGLSAAVCCSS 87
T N L EC+ SPCG A+ C++
Sbjct 22 TIENPLRTECLQFSPCGDYLALGCAN 47
Lambda K H
0.320 0.133 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1191270180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40