bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0686_orf1
Length=127
Score E
Sequences producing significant alignments: (Bits) Value
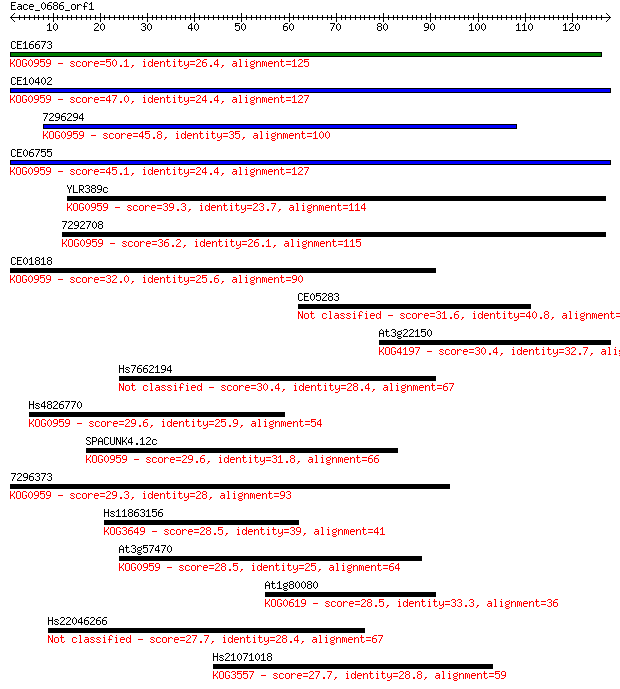
CE16673 50.1 1e-06
CE10402 47.0 8e-06
7296294 45.8 2e-05
CE06755 45.1 4e-05
YLR389c 39.3 0.002
7292708 36.2 0.013
CE01818 32.0 0.26
CE05283 31.6 0.39
At3g22150 30.4 0.73
Hs7662194 30.4 0.79
Hs4826770 29.6 1.3
SPACUNK4.12c 29.6 1.4
7296373 29.3 1.7
Hs11863156 28.5 3.0
At3g57470 28.5 3.3
At1g80080 28.5 3.5
Hs22046266 27.7 4.8
Hs21071018 27.7 5.5
> CE16673
Length=301
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/125 (26%), Positives = 56/125 (44%), Gaps = 1/125 (0%)
Query 1 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTH 60
+G+ + D + ++ + +D+LRT+ LGYI + T LQ V+G K+
Sbjct 80 IGVQNTYDNAVIGLIKNLITEPAFDTLRTKESLGYIVWTRTHFNCGTVALQILVQGPKS- 138
Query 61 PDEVVKMIDEELTKAKEYLANMPDAEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFSHS 120
D V++ I+ L ++ + MP E A+L + S FKK +EI
Sbjct 139 VDHVLERIEAFLESVRKEIVEMPQEEFENRVSGLIAQLEEKPKTLSCRFKKFWDEIECRQ 198
Query 121 NCFTK 125
FT+
Sbjct 199 YNFTR 203
> CE10402
Length=1067
Score = 47.0 bits (110), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 31/127 (24%), Positives = 55/127 (43%), Gaps = 1/127 (0%)
Query 1 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTH 60
+G+ + D + ++ Q + +++LRT LGYI T L V+G K+
Sbjct 839 IGVQNTYDNAVVGLIDQLIREPAFNTLRTNEALGYIVWTGSRLNCGTVALNVIVQGPKS- 897
Query 61 PDEVVKMIDEELTKAKEYLANMPDAEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFSHS 120
D V++ I+ L ++ +A MP E A+L + S F++ EI
Sbjct 898 VDHVLERIEVFLESVRKEIAEMPQEEFDNQVSGMIARLEEKPKTLSSRFRRFWNEIECRQ 957
Query 121 NCFTKRD 127
F +R+
Sbjct 958 YNFARRE 964
> 7296294
Length=990
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 35/103 (33%), Positives = 51/103 (49%), Gaps = 4/103 (3%)
Query 8 DRVNLAV--LTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDEVV 65
D N+ V ++Q L+ YD LRT+ QLGYI + + ++ V+ AK HP V
Sbjct 776 DHTNIMVNLVSQVLSEPCYDCLRTKEQLGYIVFSGVRKVNGANGIRIIVQSAK-HPSYVE 834
Query 66 KMIDEELTKAKEYLANMPDAEMARWKEA-AHAKLTKMEANFSE 107
I+ L + + +MP E R KEA A KL K + F +
Sbjct 835 DRIENFLQTYLQVIEDMPLDEFERHKEALAVKKLEKPKTIFQQ 877
> CE06755
Length=980
Score = 45.1 bits (105), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 31/127 (24%), Positives = 56/127 (44%), Gaps = 1/127 (0%)
Query 1 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTH 60
+G+ + D + ++ Q + ++D+LRT LGYI L FV+G K+
Sbjct 753 IGVQNKYDNAVVGLIDQLIKEPVFDTLRTNEALGYIVWTGCRFNCGAVALNIFVQGPKS- 811
Query 61 PDEVVKMIDEELTKAKEYLANMPDAEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFSHS 120
D V++ I+ L ++ + MP E + A+L + S FK+ +I
Sbjct 812 VDYVLERIEVFLESVRKEIIEMPQDEFEKKVAGMIARLEEKPKTLSNRFKRFWYQIECRQ 871
Query 121 NCFTKRD 127
F +R+
Sbjct 872 YDFARRE 878
> YLR389c
Length=988
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/114 (23%), Positives = 51/114 (44%), Gaps = 1/114 (0%)
Query 13 AVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDEVVKMIDEEL 72
+ Q ++ +D+LRT+ QLGY+ + TA ++ ++ T P + I+
Sbjct 818 GLFAQLIHEPCFDTLRTKEQLGYVVFSSSLNNHGTANIRILIQSEHTTP-YLEWRINNFY 876
Query 73 TKAKEYLANMPDAEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFSHSNCFTKR 126
+ L +MP+ + + KEA L + N +E+ + I+ FT R
Sbjct 877 ETFGQVLRDMPEEDFEKHKEALCNSLLQKFKNMAEESARYTAAIYLGDYNFTHR 930
> 7292708
Length=1077
Score = 36.2 bits (82), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 30/117 (25%), Positives = 50/117 (42%), Gaps = 2/117 (1%)
Query 12 LAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEG--AKTHPDEVVKMID 69
L +L F++ ++D LRT+ QLGY GA A V KT D V I+
Sbjct 827 LDLLMMFVDEPLFDQLRTKEQLGYHVGATVRLNYGIAGYSIMVNSQETKTTADYVEGRIE 886
Query 70 EELTKAKEYLANMPDAEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFSHSNCFTKR 126
K + L ++P E +++ + S + ++ +EI + S F +R
Sbjct 887 VFRAKMLQILRHLPQDEYEHTRDSLIKLKLVADLALSTEMSRNWDEIINESYLFDRR 943
> CE01818
Length=745
Score = 32.0 bits (71), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 23/92 (25%), Positives = 39/92 (42%), Gaps = 3/92 (3%)
Query 1 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTH 60
+G+ S + +L + + Y LRT LGY + L V+G ++
Sbjct 587 IGVQSTYNNSVNKLLNELIKNPAYTILRTNEALGYNVSTESRLNDGNVYLHVIVQGPES- 645
Query 61 PDEVVKMIDEELTKAKEYLANMP--DAEMARW 90
D V++ I+ L A+E + MP D + W
Sbjct 646 ADHVLERIEVFLESAREEIVAMPQEDFDYQVW 677
> CE05283
Length=1058
Score = 31.6 bits (70), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 29/60 (48%), Gaps = 11/60 (18%)
Query 62 DEVVKMIDEELTKAKEYLANMPD-----------AEMARWKEAAHAKLTKMEANFSEDFK 110
D V++ +EELT A+ L + P+ A M RW+ +L K +NFS D K
Sbjct 650 DPRVQLNNEELTDARNVLKSSPEKAELLGKLRKSATMQRWRATDVNELLKELSNFSNDVK 709
> At3g22150
Length=820
Score = 30.4 bits (67), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 23/49 (46%), Gaps = 1/49 (2%)
Query 79 LANMPDAEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFSHSNCFTKRD 127
L+NM AE +WK + E ++ +S EI + NCF RD
Sbjct 733 LSNMY-AEEQKWKSVDKVRRGMREKGLKKEVGRSGIEIAGYVNCFVSRD 780
> Hs7662194
Length=1462
Score = 30.4 bits (67), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 32/67 (47%), Gaps = 1/67 (1%)
Query 24 YDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDEVVKMIDEELTKAKEYLANMP 83
+D + + AG E+ A + L + E + TH EV +M+ E+L + Y+ M
Sbjct 895 HDRVHLRSTYHRYAGHLEASADCSRALS-YYEKSDTHRFEVPRMLSEDLPSLELYVNKMK 953
Query 84 DAEMARW 90
D + RW
Sbjct 954 DKTLWRW 960
> Hs4826770
Length=1019
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 14/54 (25%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 5 SIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAK 58
S + + L + Q ++ +++LRT+ QLGYI + +A L+ ++ K
Sbjct 801 STSENMFLELFCQIISEPCFNTLRTKEQLGYIVFSGPRRANGIQSLRFIIQSEK 854
> SPACUNK4.12c
Length=969
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 21/80 (26%), Positives = 31/80 (38%), Gaps = 14/80 (17%)
Query 17 QFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKT--------------HPD 62
Q + + LRT+ QLGYI Q L FV+ ++
Sbjct 774 QIMKEPTFSILRTKEQLGYIVFTLVRQVTPFINLNIFVQSERSSTYLESRIRALLDQFKS 833
Query 63 EVVKMIDEELTKAKEYLANM 82
E ++M DE+ +K K L N
Sbjct 834 EFLEMSDEDFSKHKSSLINF 853
> 7296373
Length=908
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 26/95 (27%), Positives = 39/95 (41%), Gaps = 7/95 (7%)
Query 1 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKE--SQAASTALLQCFVEGAK 58
L + I D V L V F N+ LRT+ QLGY G + L+ + K
Sbjct 667 LKMECIMDLVELIVEEPFFNQ-----LRTQEQLGYSLGIHQRIGYGVLAFLITINTQETK 721
Query 59 THPDEVVKMIDEELTKAKEYLANMPDAEMARWKEA 93
D V + I+ ++ E ++ M D E +E
Sbjct 722 HRADYVEQRIEAFRSRMAELVSQMSDTEFKNIRET 756
> Hs11863156
Length=971
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 24/44 (54%), Gaps = 3/44 (6%)
Query 21 RRIYDSLRTEAQL--GYIAGAKESQAASTALLQCFVEGAKT-HP 61
+RI S +TE ++ G I G K L QCF+E +++ HP
Sbjct 230 KRILMSKKTEMEIVDGLIEGCKTQPLPQDPLWQCFLESSQSVHP 273
> At3g57470
Length=989
Score = 28.5 bits (62), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 16/64 (25%), Positives = 26/64 (40%), Gaps = 0/64 (0%)
Query 24 YDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDEVVKMIDEELTKAKEYLANMP 83
+ LRT QLGYI S + +Q ++ + P + ++ L + NM
Sbjct 809 FHQLRTIEQLGYITSLSLSNDSGVYGVQFIIQSSVKGPGHIDSRVESLLKDLESKFYNMS 868
Query 84 DAEM 87
D E
Sbjct 869 DEEF 872
> At1g80080
Length=496
Score = 28.5 bits (62), Expect = 3.5, Method: Composition-based stats.
Identities = 12/36 (33%), Positives = 18/36 (50%), Gaps = 0/36 (0%)
Query 55 EGAKTHPDEVVKMIDEELTKAKEYLANMPDAEMARW 90
+GA+T PDE + D ++ A +PD RW
Sbjct 50 DGARTEPDEQDAVYDIMRATGNDWAAAIPDVCRGRW 85
> Hs22046266
Length=390
Score = 27.7 bits (60), Expect = 4.8, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 24/67 (35%), Gaps = 0/67 (0%)
Query 9 RVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDEVVKMI 68
R V + L R I LR A K LL CFV+ HP + +
Sbjct 317 RPQCRVKVKLLQRSISSLLRFAAGEDGSYEVKSVLGKEVGLLNCFVQSVTAHPTSCIGLE 376
Query 69 DEELTKA 75
+ EL A
Sbjct 377 EIELLSA 383
> Hs21071018
Length=594
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 17/59 (28%), Positives = 28/59 (47%), Gaps = 1/59 (1%)
Query 44 AASTALLQCFVEGAKTHPDEVVKMIDEELTKAKEYLANMPDAEMARWKEAAHAKLTKME 102
ST L QC GA+ + K ++EEL +++ L + + RW+ A + ME
Sbjct 123 GTSTLLFQCQEVGAERLKTSLQKALEEELEQSRPRLGGLQPGQ-DRWRGPAMERPLPME 180
Lambda K H
0.315 0.128 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1176738752
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40