bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0681_orf4
Length=746
Score E
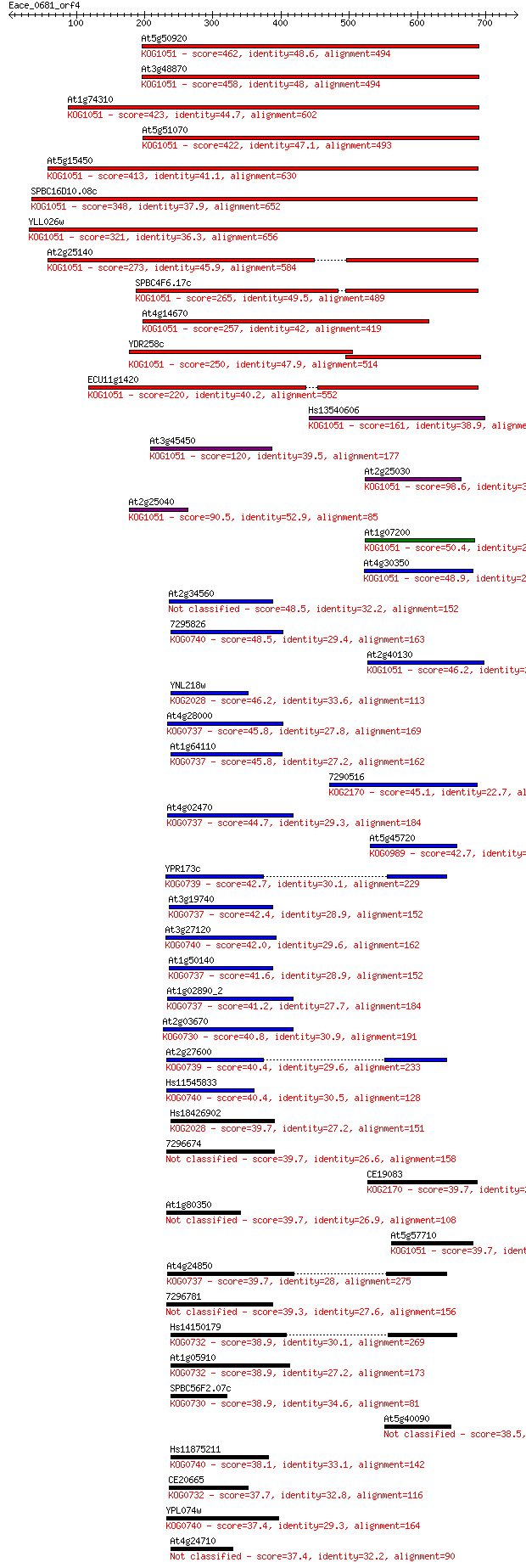
Sequences producing significant alignments: (Bits) Value
At5g50920 462 1e-129
At3g48870 458 2e-128
At1g74310 423 7e-118
At5g51070 422 1e-117
At5g15450 413 6e-115
SPBC16D10.08c 348 2e-95
YLL026w 321 4e-87
At2g25140 273 1e-72
SPBC4F6.17c 265 2e-70
At4g14670 257 5e-68
YDR258c 250 7e-66
ECU11g1420 220 1e-56
Hs13540606 161 6e-39
At3g45450 120 1e-26
At2g25030 98.6 4e-20
At2g25040 90.5 1e-17
At1g07200 50.4 2e-05
At4g30350 48.9 4e-05
At2g34560 48.5 6e-05
7295826 48.5 6e-05
At2g40130 46.2 3e-04
YNL218w 46.2 3e-04
At4g28000 45.8 3e-04
At1g64110 45.8 3e-04
7290516 45.1 7e-04
At4g02470 44.7 8e-04
At5g45720 42.7 0.003
YPR173c 42.7 0.003
At3g19740 42.4 0.004
At3g27120 42.0 0.006
At1g50140 41.6 0.006
At1g02890_2 41.2 0.008
At2g03670 40.8 0.010
At2g27600 40.4 0.014
Hs11545833 40.4 0.016
Hs18426902 39.7 0.024
7296674 39.7 0.025
CE19083 39.7 0.026
At1g80350 39.7 0.027
At5g57710 39.7 0.028
At4g24850 39.7 0.028
7296781 39.3 0.029
Hs14150179 38.9 0.040
At1g05910 38.9 0.041
SPBC56F2.07c 38.9 0.042
At5g40090 38.5 0.050
Hs11875211 38.1 0.072
CE20665 37.7 0.098
YPL074w 37.4 0.12
At4g24710 37.4 0.14
> At5g50920
Length=929
Score = 462 bits (1190), Expect = 1e-129, Method: Compositional matrix adjust.
Identities = 240/519 (46%), Positives = 349/519 (67%), Gaps = 31/519 (5%)
Query 196 TISQYGVDLSFMAQQGLLPPVIGRDEEIDRIAQILSRKMTKAPMLLGEPGVGKTAVIEGL 255
T+ +YG +L+ +A++G L PV+GR +I+R+ QIL R+ P L+GEPGVGKTA+ EGL
Sbjct 256 TLEEYGTNLTKLAEEGKLDPVVGRQPQIERVVQILGRRTKNNPCLIGEPGVGKTAIAEGL 315
Query 256 AQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYEKRMKEIISYLQAHADEVILFIDE 315
AQRI G VPE++ +++ +LD+ L AG+ RGE+E+R+K+++ ++ +DE+ILFIDE
Sbjct 316 AQRIASGDVPETIEGKKVITLDMGLLVAGTKYRGEFEERLKKLMEEIR-QSDEIILFIDE 374
Query 316 IHTLIGAGKAAGSMDASQILKVPLARGEIVLVGATTLSEYKLYIEKDAAFCRRFQKIVVE 375
+HTLIGAG A G++DA+ ILK LARGE+ +GATTL EY+ +IEKD A RRFQ + V
Sbjct 375 VHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEYRKHIEKDPALERRFQPVKVP 434
Query 376 APSKERTLGILQKVRTKYEDHHNMDIPDEVLAAVVGLSDQYIKRRSFPDKALDLLDESCA 435
P+ + T+ IL+ +R +YE HH + DE L A LS QYI R PDKA+DL+DE+ +
Sbjct 435 EPTVDETIQILKGLRERYEIHHKLRYTDESLVAAAQLSYQYISDRFLPDKAIDLIDEAGS 494
Query 436 MRRVRFNNRVAEVGKQLEDHRAHRVTLSEEE------------LKELEEEYR-------- 475
R+R + +V E ++LE ++T + E L++ E E R
Sbjct 495 RVRLR-HAQVPEEARELEKE-LRQITKEKNEAVRGQDFEKAGTLRDREIELRAEVSAIQA 552
Query 476 -----TLTEPTDDRPDPDRIELKVDDVARVLSVWTGIPMGKMTEDEMSRILKLADVLGKR 530
+ E P E D+ ++S WTGIP+ K++ DE R+LK+ + L KR
Sbjct 553 KGKEMSKAESETGEEGPMVTE---SDIQHIVSSWTGIPVEKVSTDESDRLLKMEETLHKR 609
Query 531 VIGQDEAVQSVANAIRNHRAGLTEEKKPIGAFLFLGSSGVGKTELAKALAEEVFHSEKNL 590
+IGQDEAV++++ AIR R GL +PI +F+F G +GVGK+ELAKALA F SE+ +
Sbjct 610 IIGQDEAVKAISRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKALAAYYFGSEEAM 669
Query 591 IRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSL 650
IRLDM E+ E H++S+LIG PPGY+G EGGQLTEAVR++P++VVLFDE+E AH +++++
Sbjct 670 IRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNM 729
Query 651 LLPMLDEGHLTDTKNVRVDFTSTIIIMTSNIGQQYILEG 689
+L +L++G LTD+K VDF +T++IMTSN+G I +G
Sbjct 730 MLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKG 768
> At3g48870
Length=952
Score = 458 bits (1179), Expect = 2e-128, Method: Compositional matrix adjust.
Identities = 237/519 (45%), Positives = 342/519 (65%), Gaps = 31/519 (5%)
Query 196 TISQYGVDLSFMAQQGLLPPVIGRDEEIDRIAQILSRKMTKAPMLLGEPGVGKTAVIEGL 255
T+ +YG +L+ +A++G L PV+GR +I+R+ QIL+R+ P L+GEPGVGKTA+ EGL
Sbjct 277 TLEEYGTNLTKLAEEGKLDPVVGRQPQIERMVQILARRTKNNPCLIGEPGVGKTAIAEGL 336
Query 256 AQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYEKRMKEIISYLQAHADEVILFIDE 315
AQRI G VPE++ + + +LD+ L AG+ RGE+E+R+K+++ ++ +DE+ILFIDE
Sbjct 337 AQRIASGDVPETIEGKTVITLDMGLLVAGTKYRGEFEERLKKLMEEIR-QSDEIILFIDE 395
Query 316 IHTLIGAGKAAGSMDASQILKVPLARGEIVLVGATTLSEYKLYIEKDAAFCRRFQKIVVE 375
+HTLIGAG A G++DA+ ILK LARGE+ +GATT+ EY+ +IEKD A RRFQ + V
Sbjct 396 VHTLIGAGAAEGAIDAANILKPALARGELQCIGATTIDEYRKHIEKDPALERRFQPVKVP 455
Query 376 APSKERTLGILQKVRTKYEDHHNMDIPDEVLAAVVGLSDQYIKRRSFPDKALDLLDESCA 435
P+ E + ILQ +R +YE HH + DE L A LS QYI R PDKA+DL+DE+ +
Sbjct 456 EPTVEEAIQILQGLRERYEIHHKLRYTDEALVAAAQLSHQYISDRFLPDKAIDLIDEAGS 515
Query 436 MRRVR---FNNRVAEVGKQLE-------------------DHRAHRVTLSEE---ELKEL 470
R+R E+ KQL HR + L E L
Sbjct 516 RVRLRHAQLPEEARELEKQLRQITKEKNEAVRSQDFEMAGSHRDREIELKAEIANVLSRG 575
Query 471 EEEYRTLTEPTDDRPDPDRIELKVDDVARVLSVWTGIPMGKMTEDEMSRILKLADVLGKR 530
+E + E + P + D+ +++ WTGIP+ K++ DE SR+L++ L R
Sbjct 576 KEVAKAENEAEEGGPT-----VTESDIQHIVATWTGIPVEKVSSDESSRLLQMEQTLHTR 630
Query 531 VIGQDEAVQSVANAIRNHRAGLTEEKKPIGAFLFLGSSGVGKTELAKALAEEVFHSEKNL 590
VIGQDEAV++++ AIR R GL +PI +F+F G +GVGK+ELAKALA F SE+ +
Sbjct 631 VIGQDEAVKAISRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKALAAYYFGSEEAM 690
Query 591 IRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSL 650
IRLDM E+ E H++S+LIG PPGY+G EGGQLTEAVR++P+++VLFDE+E AH +++++
Sbjct 691 IRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTLVLFDEIEKAHPDVFNM 750
Query 651 LLPMLDEGHLTDTKNVRVDFTSTIIIMTSNIGQQYILEG 689
+L +L++G LTD+K VDF +T++IMTSN+G I +G
Sbjct 751 MLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKG 789
> At1g74310
Length=911
Score = 423 bits (1088), Expect = 7e-118, Method: Compositional matrix adjust.
Identities = 269/678 (39%), Positives = 386/678 (56%), Gaps = 88/678 (12%)
Query 88 RSVKDFIVDLPRSSTPINLGESGPSVALRNALRKSWKMAESEG-SPIGVQHLYAALIDIP 146
R + + LP S P + + S +L +R++ +S G + + V L L++
Sbjct 64 RVINQALKKLPSQSPPPD--DIPASSSLIKVIRRAQAAQKSRGDTHLAVDQLIMGLLEDS 121
Query 147 AFQKIMAKSGCDLKIFKSEVVANFKGRQRGVFPTSFLDMVGDKQGASDVTISQYGVDLSF 206
+ ++ + G KSEV + RG GD + + YG DL
Sbjct 122 QIRDLLNEVGVATARVKSEV-----EKLRGKEGKKVESASGDTNFQA---LKTYGRDL-- 171
Query 207 MAQQGLLPPVIGRDEEIDRIAQILSRKMTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVPE 266
+ Q G L PVIGRDEEI R+ +ILSR+ P+L+GEPGVGKTAV+EGLAQRIV+G VP
Sbjct 172 VEQAGKLDPVIGRDEEIRRVVRILSRRTKNNPVLIGEPGVGKTAVVEGLAQRIVKGDVPN 231
Query 267 SLRNRRIFSLDLLSLSAGSAMRGEYEKRMKEIISYLQAHADEVILFIDEIHTLIGAGKAA 326
SL + R+ SLD+ +L AG+ RGE+E+R+K ++ ++ +VILFIDEIH ++GAGK
Sbjct 232 SLTDVRLISLDMGALVAGAKYRGEFEERLKSVLKEVEDAEGKVILFIDEIHLVLGAGKTE 291
Query 327 GSMDASQILKVPLARGEIVLVGATTLSEYKLYIEKDAAFCRRFQKIVVEAPSKERTLGIL 386
GSMDA+ + K LARG++ +GATTL EY+ Y+EKDAAF RRFQ++ V PS T+ IL
Sbjct 292 GSMDAANLFKPMLARGQLRCIGATTLEEYRKYVEKDAAFERRFQQVYVAEPSVPDTISIL 351
Query 387 QKVRTKYEDHHNMDIPDEVLAAVVGLSDQYIKRRSFPDKALDLLDESCAMRRVRFNNRVA 446
+ ++ KYE HH + I D L LS +YI R PDKA+DL+DE+CA RV+ +++
Sbjct 352 RGLKEKYEGHHGVRIQDRALINAAQLSARYITGRHLPDKAIDLVDEACANVRVQLDSQPE 411
Query 447 EVGK---------------QLEDHRAHRVTLSE---------EELKELEEEYRTLTEPTD 482
E+ + E +A + L E ++L+ L +YR E D
Sbjct 412 EIDNLERKRMQLEIELHALEREKDKASKARLIEVRKELDDLRDKLQPLTMKYRKEKERID 471
Query 483 D--RPDPDRIELKVD--------DVAR--------VLSVWTGIPMGKMTEDEMSRILK-- 522
+ R R EL D+AR + V + I + T E + +L
Sbjct 472 EIRRLKQKREELMFSLQEAERRYDLARAADLRYGAIQEVESAIAQLEGTSSEENVMLTEN 531
Query 523 -----LADVLGKRV------IGQDEAVQSVANAIRNH--------------------RAG 551
+A+V+ + +GQ+E + + A R H RAG
Sbjct 532 VGPEHIAEVVSRWTGIPVTRLGQNEKERLIGLADRLHKRVVGQNQAVNAVSEAILRSRAG 591
Query 552 LTEEKKPIGAFLFLGSSGVGKTELAKALAEEVFHSEKNLIRLDMVEYQEAHSISRLIGPP 611
L ++P G+FLFLG +GVGKTELAKALAE++F E L+R+DM EY E HS+SRLIG P
Sbjct 592 LGRPQQPTGSFLFLGPTGVGKTELAKALAEQLFDDENLLVRIDMSEYMEQHSVSRLIGAP 651
Query 612 PGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDTKNVRVDFT 671
PGY+G++EGGQLTEAVR++P+ V+LFDEVE AH +++ LL +LD+G LTD + VDF
Sbjct 652 PGYVGHEEGGQLTEAVRRRPYCVILFDEVEKAHVAVFNTLLQVLDDGRLTDGQGRTVDFR 711
Query 672 STIIIMTSNIGQQYILEG 689
+++IIMTSN+G +++L G
Sbjct 712 NSVIIMTSNLGAEHLLAG 729
> At5g51070
Length=945
Score = 422 bits (1086), Expect = 1e-117, Method: Compositional matrix adjust.
Identities = 232/523 (44%), Positives = 329/523 (62%), Gaps = 36/523 (6%)
Query 197 ISQYGVDLSFMAQQGLLPPVIGRDEEIDRIAQILSRKMTKAPMLLGEPGVGKTAVIEGLA 256
+ Q+ VDL+ A +GL+ PVIGR++E+ R+ QIL R+ P+LLGE GVGKTA+ EGLA
Sbjct 271 LEQFCVDLTARASEGLIDPVIGREKEVQRVIQILCRRTKNNPILLGEAGVGKTAIAEGLA 330
Query 257 QRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYEKRMKEIISYLQAHADEVILFIDEI 316
I E + P L +RI SLD+ L AG+ RGE E R+ +IS ++ + +VILFIDE+
Sbjct 331 ISIAEASAPGFLLTKRIMSLDIGLLMAGAKERGELEARVTALISEVK-KSGKVILFIDEV 389
Query 317 HTLIGAG------KAAGSMDASQILKVPLARGEIVLVGATTLSEYKLYIEKDAAFCRRFQ 370
HTLIG+G K +G +D + +LK L RGE+ + +TTL E++ EKD A RRFQ
Sbjct 390 HTLIGSGTVGRGNKGSG-LDIANLLKPSLGRGELQCIASTTLDEFRSQFEKDKALARRFQ 448
Query 371 KIVVEAPSKERTLGILQKVRTKYEDHHNMDIPDEVLAAVVGLSDQYIKRRSFPDKALDLL 430
+++ PS+E + IL +R KYE HHN E + A V LS +YI R PDKA+DL+
Sbjct 449 PVLINEPSEEDAVKILLGLREKYEAHHNCKYTMEAIDAAVYLSSRYIADRFLPDKAIDLI 508
Query 431 DESCAMRRV-----RFNNRVAEVGKQLEDH--------RAHRVTLSE-----------EE 466
DE+ + R+ + + + + K D+ H V LS +E
Sbjct 509 DEAGSRARIEAFRKKKEDAICILSKPPNDYWQEIKTVQAMHEVVLSSRQKQDDGDAISDE 568
Query 467 LKELEEEYRTLTEPTDDRPDPDRIELKVDDVARVLSVWTGIPMGKMTEDEMSRILKLADV 526
EL EE DD P I + DD+A V SVW+GIP+ ++T DE ++ L D
Sbjct 569 SGELVEESSLPPAAGDDEP----ILVGPDDIAAVASVWSGIPVQQITADERMLLMSLEDQ 624
Query 527 LGKRVIGQDEAVQSVANAIRNHRAGLTEEKKPIGAFLFLGSSGVGKTELAKALAEEVFHS 586
L RV+GQDEAV +++ A++ R GL + +PI A LF G +GVGKTEL KALA F S
Sbjct 625 LRGRVVGQDEAVAAISRAVKRSRVGLKDPDRPIAAMLFCGPTGVGKTELTKALAANYFGS 684
Query 587 EKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKN 646
E++++RLDM EY E H++S+LIG PPGY+G +EGG LTEA+R++P +VVLFDE+E AH +
Sbjct 685 EESMLRLDMSEYMERHTVSKLIGSPPGYVGFEEGGMLTEAIRRRPFTVVLFDEIEKAHPD 744
Query 647 LWSLLLPMLDEGHLTDTKNVRVDFTSTIIIMTSNIGQQYILEG 689
++++LL + ++GHLTD++ RV F + +IIMTSN+G I +G
Sbjct 745 IFNILLQLFEDGHLTDSQGRRVSFKNALIIMTSNVGSLAIAKG 787
> At5g15450
Length=968
Score = 413 bits (1062), Expect = 6e-115, Method: Compositional matrix adjust.
Identities = 259/716 (36%), Positives = 400/716 (55%), Gaps = 104/716 (14%)
Query 58 HVAICLLEGPHKEISQEVVGKCNGDSDFLLRSVKDFIVDLPRSSTPINLGESGPSVALRN 117
H+ LLE + +++ + K D+ +L + + FI P+ G++ S+ R+
Sbjct 109 HLMKALLEQKNG-LARRIFSKIGVDNTKVLEATEKFIQRQPKV-----YGDAAGSMLGRD 162
Query 118 --ALRKSWKMAESE--GSPIGVQHLYAALIDIPAFQKIMAKSGCDLKIFKSEV---VANF 170
AL + + + + S + V+HL A D F K + K D +I + + + +
Sbjct 163 LEALFQRARQFKKDLKDSYVSVEHLVLAFADDKRFGKQLFK---DFQISERSLKSAIESI 219
Query 171 KGRQRGVFPTSFLDMVGDKQGASDVTISQYGVDLSFMAQQGLLPPVIGRDEEIDRIAQIL 230
+G+Q S +D D +G + + +YG DL+ MA++G L PVIGRD+EI R QIL
Sbjct 220 RGKQ------SVIDQ--DPEGKYE-ALEKYGKDLTAMAREGKLDPVIGRDDEIRRCIQIL 270
Query 231 SRKMTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGE 290
SR+ P+L+GEPGVGKTA+ EGLAQRIV+G VP++L NR++ SLD+ +L AG+ RGE
Sbjct 271 SRRTKNNPVLIGEPGVGKTAISEGLAQRIVQGDVPQALMNRKLISLDMGALIAGAKYRGE 330
Query 291 YEKRMKEIISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPLARGEIVLVGAT 350
+E R+K ++ + ++ILFIDEIHT++GAG G+MDA +LK L RGE+ +GAT
Sbjct 331 FEDRLKAVLKEVTDSEGQIILFIDEIHTVVGAGATNGAMDAGNLLKPMLGRGELRCIGAT 390
Query 351 TLSEYKLYIEKDAAFCRRFQKIVVEAPSKERTLGILQKVRTKYEDHHNMDIPDEVLAAVV 410
TL EY+ YIEKD A RRFQ++ V+ P+ E T+ IL+ +R +YE HH + I D L
Sbjct 391 TLDEYRKYIEKDPALERRFQQVYVDQPTVEDTISILRGLRERYELHHGVRISDSALVEAA 450
Query 411 GLSDQYIKRRSFPDKALDLLDESCA------------------------MRRVRFNNRVA 446
LSD+YI R PDKA+DL+DE+ A M R+ N
Sbjct 451 ILSDRYISGRFLPDKAIDLVDEAAAKLKMEITSKPTALDELDRSVIKLEMERLSLTNDTD 510
Query 447 EVGKQLEDHRAHRVTLSEEELKELEEEYR----TLTEPTDDRPDPDRIELKVDDVARVLS 502
+ ++ + + L +E+ EL E++ ++ + + DR+ L++ R
Sbjct 511 KASRERLNRIETELVLLKEKQAELTEQWEHERSVMSRLQSIKEEIDRVNLEIQQAEREYD 570
Query 503 VWTGIPMGKMTEDEMSRILKLAD------------VLGKRVIGQDEA----------VQS 540
+ + + + + R L A+ + + V+G D A V
Sbjct 571 LNRAAELKYGSLNSLQRQLNEAEKELNEYLSSGKSMFREEVLGSDIAEIVSKWTGIPVSK 630
Query 541 VANAIRNHRAGLTEE--KKPIG---------------------------AFLFLGSSGVG 571
+ + R+ L EE K+ +G +F+F+G +GVG
Sbjct 631 LQQSERDKLLHLEEELHKRVVGQNPAVTAVAEAIQRSRAGLSDPGRPIASFMFMGPTGVG 690
Query 572 KTELAKALAEEVFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKP 631
KTELAKALA +F++E+ L+R+DM EY E H++SRLIG PPGY+G +EGGQLTE VR++P
Sbjct 691 KTELAKALASYMFNTEEALVRIDMSEYMEKHAVSRLIGAPPGYVGYEEGGQLTETVRRRP 750
Query 632 HSVVLFDEVENAHKNLWSLLLPMLDEGHLTDTKNVRVDFTSTIIIMTSNIGQQYIL 687
+SV+LFDE+E AH +++++ L +LD+G +TD++ V FT+T+IIMTSN+G Q+IL
Sbjct 751 YSVILFDEIEKAHGDVFNVFLQILDDGRVTDSQGRTVSFTNTVIIMTSNVGSQFIL 806
> SPBC16D10.08c
Length=905
Score = 348 bits (894), Expect = 2e-95, Method: Compositional matrix adjust.
Identities = 247/749 (32%), Positives = 385/749 (51%), Gaps = 115/749 (15%)
Query 35 AKLVTECVELLAWYLQNDRLAVAHVAICLLEGPHKE---ISQEVVGKCNGDSDFLLRSVK 91
AK +++ + Y + +L H+A LL + + +V K GD RSV
Sbjct 11 AKTLSDAYSIAQSY-GHSQLTPIHIAAALLSDSDSNGTTLLRTIVDKAGGDGQKFERSVT 69
Query 92 DFIVDLPRSSTPINLGESGPSVA--LRNA--LRKSWKMAESEGSPIGVQHLYAALIDIPA 147
+V LP P P A LRNA L+K+ K S I H A
Sbjct 70 SRLVRLPAQDPPPEQVTLSPESAKLLRNAHELQKTQK-----DSYIAQDHFIAVFTKDDT 124
Query 148 FQKIMAKSGCDLKIFKSEVVANFKGRQRGVFPTSFLDMVGDKQGASDVTISQYGVDLSFM 207
+ ++A++G K F+ V N +G +R +D ++G ++++ VDL+ +
Sbjct 125 LKSLLAEAGVTPKAFEF-AVNNVRGNKR-------IDSKNAEEGFD--ALNKFTVDLTEL 174
Query 208 AQQGLLPPVIGRDEEIDRIAQILSRKMTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVPES 267
A+ G L PVIGR++EI R ++LSR+ P+L+GEPGVGKT++ EGLA+RI++ VP +
Sbjct 175 ARNGQLDPVIGREDEIRRTIRVLSRRTKNNPVLIGEPGVGKTSIAEGLARRIIDDDVPAN 234
Query 268 LRNRRIFSLDLLSLSAGSAMRGEYEKRMKEIISYLQAHADEVILFIDEIHTLI-GAGKAA 326
L N ++ SLD+ SL AGS RGE+E+R+K ++ ++ +ILF+DE+H L+
Sbjct 235 LSNCKLLSLDVGSLVAGSKFRGEFEERIKSVLKEVEESETPIILFVDEMHLLMGAGSGGE 294
Query 327 GSMDASQILKVPLARGEIVLVGATTLSEYKLYIEKDAAFCRRFQKIVVEAPSKERTLGIL 386
G MDA+ +LK LARG++ +GATTL+EYK YIEKDAAF RRFQ I+V+ PS E T+ IL
Sbjct 295 GGMDAANLLKPMLARGKLHCIGATTLAEYKKYIEKDAAFERRFQIILVKEPSIEDTISIL 354
Query 387 QKVRTKYEDHHNMDIPDEVLAAVVGLSDQYIKRRSFPDKALDLLDESCA----------- 435
+ ++ KYE HH + I D L L+ +Y+ R PD A+DL+DE+ A
Sbjct 355 RGLKEKYEVHHGVTISDRALVTAAHLASRYLTSRRLPDSAIDLVDEAAAAVRVTRESQPE 414
Query 436 --------MRRVRFNNRVAEVGKQLEDHRAHRVTLSEEELKELEEEYRTL---------- 477
+R++R R E ++ ++ R+ + +E +++EEE R +
Sbjct 415 VLDNLERKLRQLRVEIRALE--REKDEASKERLKAARKEAEQVEEETRPIREKYELEKSR 472
Query 478 -TEPTDDRPDPDRIELKVDDVAR----VLSV---WTGIPMGKMTEDEMSRILKLADV--- 526
+E D + D ++ K +D R L+ + GIP + + + + + AD
Sbjct 473 GSELQDAKRRLDELKAKAEDAERRNDFTLAADLKYYGIPDLQKRIEYLEQQKRKADAEAI 532
Query 527 ---------LGKRVIGQDEAVQSVA--NAIRNHRAGLTEEKKPIGAFLFLGSSGVGKTEL 575
L V+G D+ + VA I R TE+++ + L +G+ E
Sbjct 533 ANAQPGSEPLLIDVVGPDQINEIVARWTGIPVTRLKTTEKERLLNMEKVLSKQVIGQNEA 592
Query 576 AKALAEEV--------------------------------------FHSEKNLIRLDMVE 597
A+A + F E +IR+DM E
Sbjct 593 VTAVANAIRLSRAGLSDPNQPIASFLFCGPSGTGKTLLTKALASFMFDDENAMIRIDMSE 652
Query 598 YQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDE 657
Y E HS+SRLIG PPGY+G++ GGQLTE +R++P+SV+LFDE+E A + ++LL +LD+
Sbjct 653 YMEKHSVSRLIGAPPGYVGHEAGGQLTEQLRRRPYSVILFDEIEKAAPEVLTVLLQVLDD 712
Query 658 GHLTDTKNVRVDFTSTIIIMTSNIGQQYI 686
G +T + VD + +IIMTSN+G +Y+
Sbjct 713 GRITSGQGQVVDAKNAVIIMTSNLGAEYL 741
> YLL026w
Length=908
Score = 321 bits (823), Expect = 4e-87, Method: Compositional matrix adjust.
Identities = 238/743 (32%), Positives = 374/743 (50%), Gaps = 102/743 (13%)
Query 31 LTPDAKLVTECVELLAWYLQNDRLAVAHVAICLLEGPHK---EISQEVVGKCNGDSDFLL 87
T A + + LA Q+ +L H+ +E P Q ++ K D D
Sbjct 7 FTERALTILTLAQKLASDHQHPQLQPIHILAAFIETPEDGSVPYLQNLIEKGRYDYDLFK 66
Query 88 RSVKDFIVDLPRSS-TPINLGESGPSVALRNALRKSWKMAESE-GSPIGVQHLYAALIDI 145
+ V +V +P+ P E PS AL L+ + K+ + + S I H+ AL +
Sbjct 67 KVVNRNLVRIPQQQPAP---AEITPSYALGKVLQDAAKIQKQQKDSFIAQDHILFALFND 123
Query 146 PAFQKIMAKSGCDLKIFKSEVVANFKGRQRGVFPTSFLDMVGDKQGASDVTISQYGVDLS 205
+ Q+I ++ D++ K + + +G R +D G +S+Y +D++
Sbjct 124 SSIQQIFKEAQVDIEAIKQQAL-ELRGNTR-------IDSRGADTNTPLEYLSKYAIDMT 175
Query 206 FMAQQGLLPPVIGRDEEIDRIAQILSRKMTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVP 265
A+QG L PVIGR+EEI ++L+R++ P L+GEPG+GKTA+IEG+AQRI++ VP
Sbjct 176 EQARQGKLDPVIGREEEIRSTIRVLARRIKSNPCLIGEPGIGKTAIIEGVAQRIIDDDVP 235
Query 266 ESLRNRRIFSLDLLSLSAGSAMRGEYEKRMKEIISYLQAHADEVILFIDEIHTLIGAGKA 325
L+ ++FSLDL +L+AG+ +G++E+R K ++ ++ ++LFIDEIH L+G GK
Sbjct 236 TILQGAKLFSLDLAALTAGAKYKGDFEERFKGVLKEIEESKTLIVLFIDEIHMLMGNGKD 295
Query 326 AGSMDASQILKVPLARGEIVLVGATTLSEYKLYIEKDAAFCRRFQKIVVEAPSKERTLGI 385
DA+ ILK L+RG++ ++GATT +EY+ +EKD AF RRFQKI V PS +T+ I
Sbjct 296 ----DAANILKPALSRGQLKVIGATTNNEYRSIVEKDGAFERRFQKIEVAEPSVRQTVAI 351
Query 386 LQKVRTKYEDHHNMDIPDEVLAAVVGLSDQYIKRRSFPDKALDLLDESCAMRRVRFNNRV 445
L+ ++ KYE HH + I D L L+ +Y+ R PD ALDL+D SCA V +++
Sbjct 352 LRGLQPKYEIHHGVRILDSALVTAAQLAKRYLPYRRLPDSALDLVDISCAGVAVARDSKP 411
Query 446 AEVG---KQLE-----------DHRA-----HRVTLSEEELKELEEEYRTL--------- 477
E+ +QL+ D A R+ L+ ++ L+EE L
Sbjct 412 EELDSKERQLQLIQVEIKALERDEDADSTTKDRLKLARQKEASLQEELEPLRQRYNEEKH 471
Query 478 --TEPTDDRPDPDRIELKVDDVARVLSVWTGIPMGKMT-EDEMSRILKLADVLG--KRVI 532
E T + D +E K D R T + D +I KL D + +R
Sbjct 472 GHEELTQAKKKLDELENKALDAERRYDTATAADLRYFAIPDIKKQIEKLEDQVAEEERRA 531
Query 533 GQDEAVQSV--ANAIRNHRAGLT----------EEKKPIGAFLFLGSSGVGKTELAKALA 580
G + +Q+V ++ I A LT E +K I L S VG+ + KA++
Sbjct 532 GANSMIQNVVDSDTISETAARLTGIPVKKLSESENEKLIHMERDLSSEVVGQMDAIKAVS 591
Query 581 EEVFHSEKNL-------------------------------------IRLDMVEYQEAHS 603
V S L IR+D E E ++
Sbjct 592 NAVRLSRSGLANPRQPASFLFLGLSGSGKTELAKKVAGFLFNDEDMMIRVDCSELSEKYA 651
Query 604 ISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDT 663
+S+L+G GY+G DEGG LT ++ KP+SV+LFDEVE AH ++ +++L MLD+G +T
Sbjct 652 VSKLLGTTAGYVGYDEGGFLTNQLQYKPYSVLLFDEVEKAHPDVLTVMLQMLDDGRITSG 711
Query 664 KNVRVDFTSTIIIMTSNIGQQYI 686
+ +D ++ I+IMTSN+G ++I
Sbjct 712 QGKTIDCSNCIVIMTSNLGAEFI 734
> At2g25140
Length=874
Score = 273 bits (697), Expect = 1e-72, Method: Compositional matrix adjust.
Identities = 163/395 (41%), Positives = 236/395 (59%), Gaps = 20/395 (5%)
Query 58 HVAICLLEGPHKEISQEVVGKCNGDSDFLLRSVKDFIVDLPRSS--TPINLGESGPSVAL 115
H+ LLE +++++ K D+ +L++ FI P S + LG S SV L
Sbjct 25 HLMKALLEQKDG-MARKIFTKAGIDNSSVLQATDLFISKQPTVSDASGQRLG-SSLSVIL 82
Query 116 RNALRKSWKMAESEGSPIGVQHLYAALIDIPAF-QKIMAKSGCDLKIFKSEVVANFKGRQ 174
NA R M +S + V+H A F Q+ D+++ K + + + +G Q
Sbjct 83 ENAKRHKKDMLDS---YVSVEHFLLAYYSDTRFGQEFFRDMKLDIQVLK-DAIKDVRGDQ 138
Query 175 RGVFPTSFLDMVGDKQGASDV-TISQYGVDLSFMAQQGLLPPVIGRDEEIDRIAQILSRK 233
R V D+ S + +YG DL+ MA++G L PVIGRD+EI R QIL R+
Sbjct 139 R----------VTDRNPESKYQALEKYGNDLTEMARRGKLDPVIGRDDEIRRCIQILCRR 188
Query 234 MTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYEK 293
P+++GEPGVGKTA+ EGLAQRIV G VPE L NR++ SLD+ SL AG+ RG++E+
Sbjct 189 TKNNPVIIGEPGVGKTAIAEGLAQRIVRGDVPEPLMNRKLISLDMGSLLAGAKFRGDFEE 248
Query 294 RMKEIISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPLARGEIVLVGATTLS 353
R+K ++ + A + ILFIDEIHT++GAG G+MDAS +LK L RGE+ +GATTL+
Sbjct 249 RLKAVMKEVSASNGQTILFIDEIHTVVGAGAMDGAMDASNLLKPMLGRGELRCIGATTLT 308
Query 354 EYKLYIEKDAAFCRRFQKIVVEAPSKERTLGILQKVRTKYEDHHNMDIPDEVLAAVVGLS 413
EY+ YIEKD A RRFQ+++ PS E T+ IL+ +R +YE HH + I D L + L+
Sbjct 309 EYRKYIEKDPALERRFQQVLCVQPSVEDTISILRGLRERYELHHGVTISDSALVSAAVLA 368
Query 414 DQYIKRRSFPDKALDLLDESCAMRRVRFNNRVAEV 448
D+YI R PDKA+DL+DE+ A ++ ++ E+
Sbjct 369 DRYITERFLPDKAIDLVDEAGAKLKMEITSKPTEL 403
Score = 236 bits (603), Expect = 1e-61, Method: Compositional matrix adjust.
Identities = 105/193 (54%), Positives = 152/193 (78%), Gaps = 0/193 (0%)
Query 496 DVARVLSVWTGIPMGKMTEDEMSRILKLADVLGKRVIGQDEAVQSVANAIRNHRAGLTEE 555
D+A ++S WTGIP+ + + E +++ L +VL RVIGQD AV+SVA+AIR RAGL++
Sbjct 530 DIAEIVSKWTGIPLSNLQQSEREKLVMLEEVLHHRVIGQDMAVKSVADAIRRSRAGLSDP 589
Query 556 KKPIGAFLFLGSSGVGKTELAKALAEEVFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYM 615
+PI +F+F+G +GVGKTELAKALA +F++E ++R+DM EY E HS+SRL+G PPGY+
Sbjct 590 NRPIASFMFMGPTGVGKTELAKALAGYLFNTENAIVRVDMSEYMEKHSVSRLVGAPPGYV 649
Query 616 GNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDTKNVRVDFTSTII 675
G +EGGQLTE VR++P+SVVLFDE+E AH +++++LL +LD+G +TD++ V F + ++
Sbjct 650 GYEEGGQLTEVVRRRPYSVVLFDEIEKAHPDVFNILLQLLDDGRITDSQGRTVSFKNCVV 709
Query 676 IMTSNIGQQYILE 688
IMTSNIG +ILE
Sbjct 710 IMTSNIGSHHILE 722
> SPBC4F6.17c
Length=803
Score = 265 bits (678), Expect = 2e-70, Method: Compositional matrix adjust.
Identities = 140/306 (45%), Positives = 199/306 (65%), Gaps = 13/306 (4%)
Query 187 GDKQGASDVTISQYGVDLSFMAQQGLLPPVIGRDEEIDRIAQILSRKMTKAPMLLGEPGV 246
G KQGA+ + +YG DL+ +A+QG L PVIGR+EEI R QILSR+ P L+G GV
Sbjct 87 GRKQGAA---LEEYGTDLTALAKQGKLDPVIGREEEIQRTIQILSRRTKNNPALVGPAGV 143
Query 247 GKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYEKRMKEIISYLQAHA 306
GKTA++EGLA RI+ G VPES++++R+ LDL +L +G+ RG++E+R+K ++S L+
Sbjct 144 GKTAIMEGLASRIIRGEVPESMKDKRVIVLDLGALISGAKFRGDFEERLKSVLSDLEGAE 203
Query 307 DEVILFIDEIHTLIGAGKAAGSMDASQILKVPLARGEIVLVGATTLSEYKLYIEKDAAFC 366
+VILF+DE+H L+G GKA GS+DAS +LK LARG++ GATTL EY+ YIEKDAA
Sbjct 204 GKVILFVDEMHLLLGFGKAEGSIDASNLLKPALARGKLHCCGATTLEEYRKYIEKDAALA 263
Query 367 RRFQKIVVEAPSKERTLGILQKVRTKYEDHHNMDIPDEVLAAVVGLSDQYIKRRSFPDKA 426
RRFQ ++V PS T+ IL+ ++ +YE HH + I D+ L S +YI R PDKA
Sbjct 264 RRFQAVMVNEPSVADTISILRGLKERYEVHHGVRITDDALVTAATYSARYITDRFLPDKA 323
Query 427 LDLLDESCAMRRV----------RFNNRVAEVGKQLEDHRAHRVTLSEEELKELEEEYRT 476
+DL+DE+C+ R+ R + ++ + +LE R T S E ++LE +
Sbjct 324 IDLVDEACSSLRLQQESKPDELRRLDRQIMTIQIELESLRKETDTTSVERREKLESKLTD 383
Query 477 LTEPTD 482
L E D
Sbjct 384 LKEEQD 389
Score = 224 bits (571), Expect = 5e-58, Method: Compositional matrix adjust.
Identities = 102/193 (52%), Positives = 148/193 (76%), Gaps = 0/193 (0%)
Query 495 DDVARVLSVWTGIPMGKMTEDEMSRILKLADVLGKRVIGQDEAVQSVANAIRNHRAGLTE 554
DD+A V+S TGIP + E ++L + +GK++IGQDEA++++A+A+R RAGL
Sbjct 468 DDIAVVVSRATGIPTTNLMRGERDKLLNMEQTIGKKIIGQDEALKAIADAVRLSRAGLQN 527
Query 555 EKKPIGAFLFLGSSGVGKTELAKALAEEVFHSEKNLIRLDMVEYQEAHSISRLIGPPPGY 614
+P+ +FLFLG +GVGKT L KALAE +F ++K +IR DM E+QE H+I+RLIG PPGY
Sbjct 528 TNRPLASFLFLGPTGVGKTALTKALAEFLFDTDKAMIRFDMSEFQEKHTIARLIGSPPGY 587
Query 615 MGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDTKNVRVDFTSTI 674
+G +E G+LTEAVR+KP++V+LFDE+E AH ++ +LLL +LDEG LTD++ +VDF ST+
Sbjct 588 IGYEESGELTEAVRRKPYAVLLFDELEKAHHDITNLLLQVLDEGFLTDSQGRKVDFRSTL 647
Query 675 IIMTSNIGQQYIL 687
I+MTSN+G ++
Sbjct 648 IVMTSNLGSDILV 660
> At4g14670
Length=623
Score = 257 bits (657), Expect = 5e-68, Method: Compositional matrix adjust.
Identities = 176/495 (35%), Positives = 262/495 (52%), Gaps = 79/495 (15%)
Query 197 ISQYGVDLSFMAQQGLLPPVIGRDEEIDRIAQILSRKMTKAPMLLGEPGVGKTAVIEGLA 256
+ YG DL + Q G L PVIGR EI R+ ++LSR+ P+L+GEPGVGKTAV+EGLA
Sbjct 129 LKTYGTDL--VEQAGKLDPVIGRHREIRRVIEVLSRRTKNNPVLIGEPGVGKTAVVEGLA 186
Query 257 QRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYEKRMKEIISYLQAHADEVILFIDEI 316
QRI++G VP +L ++ SL+ ++ AG+ +RG++E+R+K ++ ++ +V+LFIDEI
Sbjct 187 QRILKGDVPINLTGVKLISLEFGAMVAGTTLRGQFEERLKSVLKAVEEAQGKVVLFIDEI 246
Query 317 HTLIGAGKAAGSMDASQILKVPLARGEIVLVGATTLSEYKLYIEKDAAFCRRFQKIVVEA 376
H +GA KA+GS DA+++LK LARG++ +GATTL EY+ ++EKDAAF RRFQ++ V
Sbjct 247 HMALGACKASGSTDAAKLLKPMLARGQLRFIGATTLEEYRTHVEKDAAFERRFQQVFVAE 306
Query 377 PSKERTLGILQKVRTKYEDHHNMDIPDEVLAAVVGLSDQYIKRRSFPDKALDLLDESCAM 436
PS T+ IL+ ++ KYE HH + I D L LS++YI R PDKA+DL+DESCA
Sbjct 307 PSVPDTISILRGLKEKYEGHHGVRIQDRALVLSAQLSERYITGRRLPDKAIDLVDESCAH 366
Query 437 RRVRFNNRVAEVGK------QLE----------DHRAHRVTLSE--EELKELEEEYRTLT 478
+ + + + E+ QLE D +A LSE +EL +L ++ LT
Sbjct 367 VKAQLDIQPEEIDSLERKVMQLEIEIHALEKEKDDKASEARLSEVRKELDDLRDKLEPLT 426
Query 479 -----------EPTDDRPDPDRIELKVDDVARVLSVWTG--IPMGKMTEDEMSRILKLA- 524
E + + D + + + + R V + G + E E S I KL
Sbjct 427 IKYKKEKKIINETRRLKQNRDDLMIALQEAERQHDVPKAAVLKYGAIQEVE-SAIAKLEK 485
Query 525 ----DVLGKRVIGQDEAVQSVA--NAIRNHRAGLTEEKKPIGAFLFLGSSGVGKTELAKA 578
+V+ +G + + V+ I R E+K+ I L VG+ E KA
Sbjct 486 SAKDNVMLTETVGPENIAEVVSRWTGIPVTRLDQNEKKRLISLADKLHERVVGQDEAVKA 545
Query 579 LAEEVFHS--------------------------------------EKNLIRLDMVEYQE 600
+A + S E L+RLDM EY +
Sbjct 546 VAAAILRSRVGLGRPQQPSGSFLFLGPTGVGKTELAKALAEQLFDSENLLVRLDMSEYND 605
Query 601 AHSISRLIGPPPGYM 615
S+++LIG PPGY+
Sbjct 606 KFSVNKLIGAPPGYV 620
> YDR258c
Length=811
Score = 250 bits (639), Expect = 7e-66, Method: Compositional matrix adjust.
Identities = 141/328 (42%), Positives = 212/328 (64%), Gaps = 9/328 (2%)
Query 178 FPTSFLDMVGD-KQGASDVTISQYGVDLSFMAQQGLLPPVIGRDEEIDRIAQILSRKMTK 236
F +++ M D Q + Q+G +L+ +A+ G L PVIGRDEEI R QILSR+
Sbjct 78 FKRTYVQMRMDPNQQPEKPALEQFGTNLTKLARDGKLDPVIGRDEEIARAIQILSRRTKN 137
Query 237 APMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYEKRMK 296
P L+G GVGKTA+I+GLAQRIV G VP+SL+++ + +LDL SL AG+ RGE+E+R+K
Sbjct 138 NPCLIGRAGVGKTALIDGLAQRIVAGEVPDSLKDKDLVALDLGSLIAGAKYRGEFEERLK 197
Query 297 EIISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPLARGEIVLVGATTLSEYK 356
+++ + +VI+FIDE+H L+G GK GSMDAS ILK LARG + + ATTL E+K
Sbjct 198 KVLEEIDKANGKVIVFIDEVHMLLGLGKTDGSMDASNILKPKLARG-LRCISATTLDEFK 256
Query 357 LYIEKDAAFCRRFQKIVVEAPSKERTLGILQKVRTKYEDHHNMDIPDEVLAAVVGLSDQY 416
+ IEKD A RRFQ I++ PS T+ IL+ ++ +YE HH + I D L + LS++Y
Sbjct 257 I-IEKDPALSRRFQPILLNEPSVSDTISILRGLKERYEVHHGVRITDTALVSAAVLSNRY 315
Query 417 IKRRSFPDKALDLLDESCAMRRVRFNNRVAEVGKQLEDHRAHRVTLSEEELKELEEEYRT 476
I R PDKA+DL+DE+CA+ R++ ++ E+ K D ++ + EL+ L++E
Sbjct 316 ITDRFLPDKAIDLVDEACAVLRLQHESKPDEIQKL--DRAIMKIQI---ELESLKKETDP 370
Query 477 LTEPTDDRPDPDRIELKVDDVARVLSVW 504
++ + + D +E+K D++ R+ +W
Sbjct 371 VSVERREALEKD-LEMKNDELNRLTKIW 397
Score = 230 bits (587), Expect = 7e-60, Method: Compositional matrix adjust.
Identities = 108/197 (54%), Positives = 149/197 (75%), Gaps = 0/197 (0%)
Query 495 DDVARVLSVWTGIPMGKMTEDEMSRILKLADVLGKRVIGQDEAVQSVANAIRNHRAGLTE 554
DD+++V++ TGIP + + + R+L + + L +RV+GQDEA+ ++++A+R RAGLT
Sbjct 470 DDISKVVAKMTGIPTETVMKGDKDRLLYMENSLKERVVGQDEAIAAISDAVRLQRAGLTS 529
Query 555 EKKPIGAFLFLGSSGVGKTELAKALAEEVFHSEKNLIRLDMVEYQEAHSISRLIGPPPGY 614
EK+PI +F+FLG +G GKTEL KALAE +F E N+IR DM E+QE H++SRLIG PPGY
Sbjct 530 EKRPIASFMFLGPTGTGKTELTKALAEFLFDDESNVIRFDMSEFQEKHTVSRLIGAPPGY 589
Query 615 MGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDTKNVRVDFTSTI 674
+ ++ GGQLTEAVR+KP++VVLFDE E AH ++ LLL +LDEG LTD+ VDF +TI
Sbjct 590 VLSESGGQLTEAVRRKPYAVVLFDEFEKAHPDVSKLLLQVLDEGKLTDSLGHHVDFRNTI 649
Query 675 IIMTSNIGQQYILEGYK 691
I+MTSNIGQ +L K
Sbjct 650 IVMTSNIGQDILLNDTK 666
> ECU11g1420
Length=851
Score = 220 bits (560), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 128/322 (39%), Positives = 194/322 (60%), Gaps = 22/322 (6%)
Query 118 ALRKSWKMAES--EGSPIGVQHLYAALIDIPAFQKIMAKSGCDLKIFKSEVVANFKGRQR 175
L K AES EG + V L +L+++ + ++++A S D++ E N +
Sbjct 83 TLAKVLMAAESKGEGEYVSVNALVLSLLEVESIRELIANSE-DIRKKVEEQTKNKRF--- 138
Query 176 GVFPTSFLDMVGDKQGASDVT--ISQYGVDLSFMAQQGLLPPVIGRDEEIDRIAQILSRK 233
D + A D T +S++ VD+ A+Q + PVIGRDEEI R+ +ILS+K
Sbjct 139 ------------DSRNADDGTDVMSRFAVDMVEQARQNVFDPVIGRDEEIRRVLEILSKK 186
Query 234 MTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYEK 293
+L+G+PGVGKTA++ G+AQ I G P +LR RI+++D+ S+ AG++ RG++E+
Sbjct 187 TKSNAILVGKPGVGKTAIVNGIAQLIANGEAP-TLRGARIYNVDVGSMVAGTSHRGDFEE 245
Query 294 RMKEIISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPLARGEIVLVGATTLS 353
R+K ++ + VILFIDEIH ++GAGK G+MDA+ +LK LA G I +GATT
Sbjct 246 RLKTLVKEAETTPG-VILFIDEIHIVLGAGKTDGAMDAANMLKPGLAAGTIKCIGATTHD 304
Query 354 EYKLYIEKDAAFCRRFQKIVVEAPSKERTLGILQKVRTKYEDHHNMDIPDEVLAAVVGLS 413
EY+ YIE D AF RRF ++VV PS E ++ +L+ ++ + E +H + I D + V S
Sbjct 305 EYRKYIENDPAFERRFVQVVVNEPSIEDSITMLRGLKGRLEAYHGVKIADSAIVYAVNSS 364
Query 414 DQYIKRRSFPDKALDLLDESCA 435
+YI R PD A+DLLD +CA
Sbjct 365 KKYISNRRLPDVAIDLLDTACA 386
Score = 191 bits (486), Expect = 4e-48, Method: Compositional matrix adjust.
Identities = 94/234 (40%), Positives = 152/234 (64%), Gaps = 14/234 (5%)
Query 454 DHRAHRVTLSEEELKELEEEYRTLTEPTDDRPDPDRIELKVDDVARVLSVWTGIPMGKMT 513
D + + + + E ELK LE E + P+ VA ++S WTGI + ++T
Sbjct 491 DLKTNVIPVIESELKRLELESVVVILPSH--------------VAEIISRWTGIDVKRLT 536
Query 514 EDEMSRILKLADVLGKRVIGQDEAVQSVANAIRNHRAGLTEEKKPIGAFLFLGSSGVGKT 573
E R+++++ + KR+ GQD AV ++ ++I R GL ++ +P+G+FL LG +GVGKT
Sbjct 537 IKENERLMEMSSRIKKRIFGQDHAVDAIVDSILQSRVGLDDDDRPVGSFLLLGPTGVGKT 596
Query 574 ELAKALAEEVFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHS 633
ELAKA+A E+F +EK+++ +DM EY I++LIG GY+G ++GG LTE ++ +P++
Sbjct 597 ELAKAVAMELFDNEKDMLVIDMSEYGNEMGITKLIGANAGYVGYNQGGTLTEPIKGRPYN 656
Query 634 VVLFDEVENAHKNLWSLLLPMLDEGHLTDTKNVRVDFTSTIIIMTSNIGQQYIL 687
V+L DEV+ AH+ + + L +LDEG +TD K VDF + ++IMTSN+GQ+ I+
Sbjct 657 VILLDEVDLAHQTVLNTLYQLLDEGRVTDGKGAVVDFRNCVVIMTSNLGQEIIM 710
> Hs13540606
Length=707
Score = 161 bits (407), Expect = 6e-39, Method: Compositional matrix adjust.
Identities = 100/284 (35%), Positives = 157/284 (55%), Gaps = 47/284 (16%)
Query 441 FNNRVAEVGKQLEDHRAHRVTLSEEELKELEEEYRTLTEPTDDRPDPDRIELKVDDVARV 500
FNNR+ ++RA + L ++YRT+ E D +P L+ +++
Sbjct 254 FNNRL--------NNRASFKGCTALHYAVLADDYRTVKELLDGGANP----LQRNEMGHT 301
Query 501 LSVWTGIPMGKMTEDEMSRILKLADV----------------------LGKRVIGQDEAV 538
P+ E E+ ++L+ ++ L + +IGQ+ A+
Sbjct 302 -------PLDYAREGEVMKLLRTSEAKYQEKQRKREAEERRRFPLEQRLKEHIIGQESAI 354
Query 539 QSVANAIRNHRAGLTEEKKPIGAFLFLGSSGVGKTELAKALAEEVFH-SEKNLIRLDMVE 597
+V AIR G +E+ P+ FLFLGSSG+GKTELAK A+ + ++K IRLDM E
Sbjct 355 ATVGAAIRRKENGWYDEEHPL-VFLFLGSSGIGKTELAKQTAKYMHKDAKKGFIRLDMSE 413
Query 598 YQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDE 657
+QE H +++ IG PPGY+G++EGGQLT+ ++Q P++VVLFDEV+ AH ++ +++L + DE
Sbjct 414 FQERHEVAKFIGSPPGYVGHEEGGQLTKKLKQCPNAVVLFDEVDKAHPDVLTIMLQLFDE 473
Query 658 GHLTDTKNVRVDFTSTIIIMTSNIGQ----QYILEGYKEARAMA 697
G LTD K +D I IMTSN+ Q+ L+ +EA M+
Sbjct 474 GRLTDGKGKTIDCKDAIFIMTSNVASDEIAQHALQLRQEALEMS 517
> At3g45450
Length=341
Score = 120 bits (300), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 70/178 (39%), Positives = 106/178 (59%), Gaps = 18/178 (10%)
Query 209 QQGLLPPVIGRDEEIDRIAQILSRKMTKA-PMLLGEPGVGKTAVIEGLAQRIVEGAVPES 267
++G L PV+GR +I R+ QIL+R+ + L+G+PGVGK A+ EG+AQRI G VPE+
Sbjct 149 RRGKLDPVVGRQPQIKRVVQILARRTCRNNACLIGKPGVGKRAIAEGIAQRIASGDVPET 208
Query 268 LRNRRIFSLDLLSLSAGSAMRGEYEKRMKEIISYLQAHADEVILFIDEIHTLIGAGKAAG 327
++ + +++ AG+ E R + I + +D++ILFIDE+H LIGAG G
Sbjct 209 IKGK----MNV----AGNCGWNEIRWRSRGKIEEVYGQSDDIILFIDEMHLLIGAGAVEG 260
Query 328 SMDASQILKVPLARGEIVLVGATTLSEYKLYIEKDAAFCRRFQKIVVEAPSKERTLGI 385
++DA+ ILK L R E+ +Y+ +IE D A RRFQ + V P+ E + I
Sbjct 261 AIDAANILKPALERCEL---------QYRKHIENDPALERRFQPVKVPEPTVEEAIQI 309
> At2g25030
Length=265
Score = 98.6 bits (244), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 50/141 (35%), Positives = 82/141 (58%), Gaps = 33/141 (23%)
Query 523 LADVLGKRVIGQDEAVQSVANAIRNHRAGLTEEKKPIGAFLFLGSSGVGKTELAKALAEE 582
L +L +R+I QD V+SVA+AIR +AG+++ + I +F+F+G V
Sbjct 2 LEQILHERIIAQDLDVESVADAIRCSKAGISDPNRLIASFMFMGQPSV------------ 49
Query 583 VFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVEN 642
+S+L+G PGY+G +GG+LTE VR++P+SVV FDE+E
Sbjct 50 ---------------------VSQLVGASPGYVGYGDGGKLTEVVRRRPYSVVQFDEIEK 88
Query 643 AHKNLWSLLLPMLDEGHLTDT 663
H +++S+LL +LD+G +T++
Sbjct 89 PHLDVFSILLQLLDDGRITNS 109
> At2g25040
Length=135
Score = 90.5 bits (223), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 45/88 (51%), Positives = 61/88 (69%), Gaps = 5/88 (5%)
Query 178 FPTSFLDMVGDKQGASDV---TISQYGVDLSFMAQQGLLPPVIGRDEEIDRIAQILSRKM 234
FP+ L DK +D + YG DL+ MA+QG LPP+IGRD+E++R QIL R
Sbjct 49 FPS--LRRFSDKANQTDAKHEALETYGSDLTKMARQGKLPPLIGRDDEVNRCIQILCRMT 106
Query 235 TKAPMLLGEPGVGKTAVIEGLAQRIVEG 262
P+++GEPGVGKTA++EGLA+RIV+G
Sbjct 107 ESNPVIIGEPGVGKTAIVEGLAERIVKG 134
> At1g07200
Length=979
Score = 50.4 bits (119), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 44/162 (27%), Positives = 73/162 (45%), Gaps = 9/162 (5%)
Query 523 LADVLGKRVIGQDEAVQSVANAIRNHRAGLTEEKKPIGAFL-FLGSSGVGKTELAKALAE 581
L ++L ++V Q EAV +++ I + T + G +L LG VGK ++A L+E
Sbjct 621 LREILSRKVAWQTEAVNAISQIICGCKTDSTRRNQASGIWLALLGPDKVGKKKVAMTLSE 680
Query 582 EVFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVE 641
F + N I +D E S+ + G +T + +KPHSVVL + VE
Sbjct 681 VFFGGKVNYICVDF--GAEHCSLDD------KFRGKTVVDYVTGELSRKPHSVVLLENVE 732
Query 642 NAHKNLWSLLLPMLDEGHLTDTKNVRVDFTSTIIIMTSNIGQ 683
A L + G + D + + I+++TS I +
Sbjct 733 KAEFPDQMRLSEAVSTGKIRDLHGRVISMKNVIVVVTSGIAK 774
> At4g30350
Length=924
Score = 48.9 bits (115), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 46/159 (28%), Positives = 75/159 (47%), Gaps = 10/159 (6%)
Query 522 KLADVLGKRVIGQDEAVQSVANAIRNHRAGLTEEKKPIGAFLFLGSSGVGKTELAKALAE 581
KL L K V Q +A SVA AI + G + K I +F G GK+++A AL++
Sbjct 572 KLLKGLAKSVWWQHDAASSVAAAITECKHGNGKSKGDIW-LMFTGPDRAGKSKMASALSD 630
Query 582 EVFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVE 641
V S+ I L S SR+ G + EAVR+ P +V++ ++++
Sbjct 631 LVSGSQPITISLG--------SSSRM-DDGLNIRGKTALDRFAEAVRRNPFAVIVLEDID 681
Query 642 NAHKNLWSLLLPMLDEGHLTDTKNVRVDFTSTIIIMTSN 680
A L + + ++ G + D+ V + III+T+N
Sbjct 682 EADILLRNNVKIAIERGRICDSYGREVSLGNVIIILTAN 720
> At2g34560
Length=393
Score = 48.5 bits (114), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 49/163 (30%), Positives = 71/163 (43%), Gaps = 28/163 (17%)
Query 236 KAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYEKRM 295
K +L G PG GKT + + +A N F++ + S S RG+ EK +
Sbjct 146 KGILLFGPPGTGKTMLAKAVATEC----------NTTFFNIS--ASSVVSKWRGDSEKLI 193
Query 296 KEIISYLQAHADEVILFIDEIHTLIG--AGKAAGSMDASQILKVPL--------ARGEIV 345
+ + + HA I F+DEI +I G+ +AS+ LK L E+V
Sbjct 194 RVLFDLARHHAPSTI-FLDEIDAIISQRGGEGRSEHEASRRLKTELLIQMDGLQKTNELV 252
Query 346 LVGATTLSEYKLYIEKDAAFCRRFQK-IVVEAPSKERTLGILQ 387
V A T L E DAA RR +K I+V P E G+ +
Sbjct 253 FVLAAT----NLPWELDAAMLRRLEKRILVPLPDPEARRGMFE 291
> 7295826
Length=477
Score = 48.5 bits (114), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 48/172 (27%), Positives = 86/172 (50%), Gaps = 28/172 (16%)
Query 239 MLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYEKRMKEI 298
+L G PG GKT + + +A S + FS++ SL+ S G+ EK +K +
Sbjct 241 LLFGPPGTGKTLIAKSIA----------SQAKAKFFSINPSSLT--SKWVGDAEKLVKTL 288
Query 299 ISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPLARGEIVLVGATTLSEYKLY 358
+ AH I+FIDE+ +L+ + ++A +++ LK I L GA + E ++
Sbjct 289 FAVAAAH-QPAIIFIDEVDSLL-SKRSANENESTLRLKNEFL---IHLDGAASNEEIRVL 343
Query 359 I--------EKDAAFCRRF-QKIVVEAPSKERTLGILQKVRTKYEDHHNMDI 401
+ E D A RRF +++ V P++E I++K+ ++ HN+D+
Sbjct 344 VIGATNRPQELDEAVRRRFVRRLYVPLPTREARQKIIEKL--IHQVKHNLDV 393
> At2g40130
Length=907
Score = 46.2 bits (108), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 40/170 (23%), Positives = 69/170 (40%), Gaps = 6/170 (3%)
Query 527 LGKRVIGQDEAVQSVANAIRNHRAGLTEEKKPIGAFLFLGSSGVGKTELAKALAEEVFHS 586
L V GQDEA + ++ A+ +T + +G VGK ++ LAE V+ S
Sbjct 541 LTDMVSGQDEAARVISCALSQPPKSVTRRDVWLN---LVGPDTVGKRRMSLVLAEIVYQS 597
Query 587 EKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKN 646
E + +D+ ++ P G + E + + P VV + +E A +
Sbjct 598 EHRFMAVDLGAAEQGMGG---CDDPMRLRGKTMVDHIFEVMCRNPFCVVFLENIEKADEK 654
Query 647 LWSLLLPMLDEGHLTDTKNVRVDFTSTIIIMTSNIGQQYILEGYKEARAM 696
L L ++ G D+ V +TI +MTS+ Y E + +
Sbjct 655 LQMSLSKAIETGKFMDSHGREVGIGNTIFVMTSSSQGSATTTSYSEEKLL 704
> YNL218w
Length=587
Score = 46.2 bits (108), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 38/115 (33%), Positives = 55/115 (47%), Gaps = 16/115 (13%)
Query 239 MLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSA-MRGEYEKRMKE 297
+L G PGVGKT++ L + + ES R F ++ + A + +RG +EK KE
Sbjct 174 ILWGPPGVGKTSLAR-LLTKTATTSSNESNVGSRYFMIETSATKANTQELRGIFEKSKKE 232
Query 298 IISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVP-LARGEIVLVGATT 351
Q +LFIDEIH Q L +P + G+I+L+GATT
Sbjct 233 ----YQLTKRRTVLFIDEIHRF---------NKVQQDLLLPHVENGDIILIGATT 274
> At4g28000
Length=726
Score = 45.8 bits (107), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 47/179 (26%), Positives = 78/179 (43%), Gaps = 28/179 (15%)
Query 233 KMTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYE 292
K + +L G PG GKT + + +A + S+ + S GE E
Sbjct 446 KPCRGILLFGPPGTGKTMMAKAIANEAGASFINVSMS------------TITSKWFGEDE 493
Query 293 KRMKEIISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPLAR---------GE 343
K ++ + + L A I+F+DE+ +++G G +A + +K G+
Sbjct 494 KNVRALFT-LAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLMSNAGD 552
Query 344 IVLVGATTLSEYKLYIEKDAAFCRRFQ-KIVVEAPSKERTLGILQKVRTKYEDHHNMDI 401
+LV A T + L D A RRF+ +I+V PS E IL+ + +K E N+D
Sbjct 553 RILVLAATNRPFDL----DEAIIRRFERRIMVGLPSVESREKILRTLLSK-EKTENLDF 606
> At1g64110
Length=821
Score = 45.8 bits (107), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 44/172 (25%), Positives = 76/172 (44%), Gaps = 27/172 (15%)
Query 239 MLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYEKRMKEI 298
+L G PG GKT + + +A+ + S+ + S GE EK ++ +
Sbjct 551 LLFGPPGTGKTMLAKAIAKEAGASFINVSMS------------TITSKWFGEDEKNVRAL 598
Query 299 ISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPLAR---------GEIVLVGA 349
+ L + I+F+DE+ +++G G +A + +K GE +LV A
Sbjct 599 FT-LASKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSHWDGLMTKPGERILVLA 657
Query 350 TTLSEYKLYIEKDAAFCRRFQ-KIVVEAPSKERTLGILQKVRTKYEDHHNMD 400
T + L D A RRF+ +I+V P+ E IL+ + K + N+D
Sbjct 658 ATNRPFDL----DEAIIRRFERRIMVGLPAVENREKILRTLLAKEKVDENLD 705
> 7290516
Length=339
Score = 45.1 bits (105), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 49/223 (21%), Positives = 85/223 (38%), Gaps = 53/223 (23%)
Query 471 EEEYRTLTEPTDDRPDPDRIELKVDDVARVLSVWTGIPMGKMTEDEMSRILKLADVLGKR 530
E Y E DDR P RI DE+ R L+ +
Sbjct 43 EHTYCRFAECCDDRNIPARI------------------------DELERSLE------RT 72
Query 531 VIGQDEAVQSVANAIRNHRAGLTEEKKPIGAFLFLGSSGVGKTELAKALAEEVFHSEKNL 590
+IGQ Q + A++ H A + +KP+ F G G GK +A+ +A+ ++
Sbjct 73 LIGQHIVRQHIVPALKAHIASGNKSRKPL-VISFHGQPGTGKNFVAEQIADAMY------ 125
Query 591 IRLDMVEYQEAHSISRLIG----PPPGYMGNDE---GGQLTEAVRQKPHSVVLFDEVENA 643
++ ++ +++ +G P + N + + +R P S+ +FDEV+
Sbjct 126 -----LKGSRSNYVTKYLGQADFPKESEVSNYRVKINNAVRDTLRSCPRSLFIFDEVDKM 180
Query 644 HKNLWSLLLPMLDEGHLTDTKNVRVDFTSTIIIMTSNIGQQYI 686
++ L ++D D D T I I SN +I
Sbjct 181 PSGVFDQLTSLVDYNAFVDG----TDNTKAIFIFLSNTAGSHI 219
> At4g02470
Length=371
Score = 44.7 bits (104), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 54/194 (27%), Positives = 83/194 (42%), Gaps = 32/194 (16%)
Query 233 KMTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYE 292
K TK +L G PG GKT + + +A GA ++ + S++ S GE E
Sbjct 102 KPTKGILLFGPPGTGKTMLAKAVATE--AGA--------NFINISMSSIT--SKWFGEGE 149
Query 293 KRMKEIISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPLARG---------E 343
K +K + S A VI F+DE+ +++G + G +A + +K E
Sbjct 150 KYVKAVFSLASKIAPSVI-FVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDRE 208
Query 344 IVLVGATTLSEYKLYIEKDAAFCRRF-QKIVVEAPSKERTLGILQKVRTKYEDHHNMDIP 402
VLV A T + L D A RR ++++V P IL + K E P
Sbjct 209 RVLVLAATNRPFDL----DEAVIRRLPRRLMVNLPDATNRSKILSVILAKEEIA-----P 259
Query 403 DEVLAAVVGLSDQY 416
D L A+ ++D Y
Sbjct 260 DVDLEAIANMTDGY 273
> At5g45720
Length=900
Score = 42.7 bits (99), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 33/132 (25%), Positives = 61/132 (46%), Gaps = 14/132 (10%)
Query 531 VIGQDEAVQSVANAIRNHRAGLTEEKKPIGAFLFLGSSGVGKTELAKALAEEV-FHSEKN 589
++GQ+ VQ+++NAI R GL ++F G +G GKT A+ A + HS +
Sbjct 357 LLGQNLVVQALSNAIAKRRVGLL--------YVFHGPNGTGKTSCARVFARALNCHSTEQ 408
Query 590 L----IRLDMVEYQEAHS-ISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAH 644
+ V Y + + R +GP + + + +QK V++FD+ +
Sbjct 409 SKPCGVCSSCVSYDDGKNRYIREMGPVKSFDFENLLDKTNIRQQQKQQLVLIFDDCDTMS 468
Query 645 KNLWSLLLPMLD 656
+ W+ L ++D
Sbjct 469 TDCWNTLSKIVD 480
> YPR173c
Length=437
Score = 42.7 bits (99), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 44/153 (28%), Positives = 73/153 (47%), Gaps = 30/153 (19%)
Query 231 SRKMTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGE 290
+RK T +L G PG GK+ + + +A N FS+ L S GE
Sbjct 162 NRKPTSGILLYGPPGTGKSYLAKAVATEA----------NSTFFSVSSSDLV--SKWMGE 209
Query 291 YEKRMKEIISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPL----------A 340
EK +K++ + + + +I FIDE+ L G + G +AS+ +K L +
Sbjct 210 SEKLVKQLFAMARENKPSII-FIDEVDALTGT-RGEGESEASRRIKTELLVQMNGVGNDS 267
Query 341 RGEIVLVGATTLSEYKLYIEKDAAFCRRFQKIV 373
+G +VL GAT + ++L D+A RRF++ +
Sbjct 268 QGVLVL-GATNIP-WQL----DSAIRRRFERRI 294
Score = 32.7 bits (73), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 37/88 (42%), Gaps = 17/88 (19%)
Query 556 KKPIGAFLFLGSSGVGKTELAKALAEEVFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYM 615
+KP L G G GK+ LAKA+A E + ++ D+V +M
Sbjct 163 RKPTSGILLYGPPGTGKSYLAKAVATEANSTFFSVSSSDLVSK---------------WM 207
Query 616 GNDEG--GQLTEAVRQKPHSVVLFDEVE 641
G E QL R+ S++ DEV+
Sbjct 208 GESEKLVKQLFAMARENKPSIIFIDEVD 235
> At3g19740
Length=439
Score = 42.4 bits (98), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 44/165 (26%), Positives = 73/165 (44%), Gaps = 33/165 (20%)
Query 236 KAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAG---SAMRGEYE 292
K +L G PG GKT + + LA GA + +S++ S G+ E
Sbjct 186 KGILLFGPPGTGKTLLAKALATE--AGA-------------NFISITGSTLTSKWFGDAE 230
Query 293 KRMKEIISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPLARG---------- 342
K K + S+ A VI+F+DE+ +L+GA A +A++ ++
Sbjct 231 KLTKALFSFASKLA-PVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKDSQ 289
Query 343 EIVLVGATTLSEYKLYIEKDAAFCRRFQKIVVEAPSKERTLGILQ 387
I+++GAT + + DA R ++I V+ P E L IL+
Sbjct 290 RILILGATN----RPFDLDDAVIRRLPRRIYVDLPDAENRLKILK 330
> At3g27120
Length=327
Score = 42.0 bits (97), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 48/172 (27%), Positives = 73/172 (42%), Gaps = 28/172 (16%)
Query 231 SRKMTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGE 290
R K +L G PG GKT + + +A G + F + SL+ S GE
Sbjct 79 CRSPGKGLLLFGPPGTGKTMIGKAIA-----GEAKATF-----FYISASSLT--SKWIGE 126
Query 291 YEKRMKEIISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPL---------AR 341
EK ++ + VI F+DEI +L+ K+ G ++S+ LK
Sbjct 127 GEKLVRALFGVASCRQPAVI-FVDEIDSLLSQRKSDGEHESSRRLKTQFLIEMEGFDSGS 185
Query 342 GEIVLVGATTLSEYKLYIEKDAAFCRRFQK-IVVEAPSKERTLGILQKVRTK 392
+I+L+GAT + E D A RR K + + PS E I+Q + K
Sbjct 186 EQILLIGATNRPQ-----ELDEAARRRLTKRLYIPLPSSEARAWIIQNLLKK 232
> At1g50140
Length=640
Score = 41.6 bits (96), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 44/165 (26%), Positives = 73/165 (44%), Gaps = 33/165 (20%)
Query 236 KAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAG---SAMRGEYE 292
K +L G PG GKT + + LA GA + +S++ S G+ E
Sbjct 387 KGILLFGPPGTGKTLLAKALATEA--GA-------------NFISITGSTLTSKWFGDAE 431
Query 293 KRMKEIISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPLARG---------- 342
K K + S+ A VI+F+DEI +L+GA + +A++ ++
Sbjct 432 KLTKALFSFATKLA-PVIIFVDEIDSLLGARGGSSEHEATRRMRNEFMAAWDGLRSKDSQ 490
Query 343 EIVLVGATTLSEYKLYIEKDAAFCRRFQKIVVEAPSKERTLGILQ 387
I+++GAT + + DA R ++I V+ P E L IL+
Sbjct 491 RILILGATN----RPFDLDDAVIRRLPRRIYVDLPDAENRLKILK 531
> At1g02890_2
Length=563
Score = 41.2 bits (95), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 51/194 (26%), Positives = 85/194 (43%), Gaps = 36/194 (18%)
Query 233 KMTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYE 292
K TK +L G PG GKT + + +A GA + +++S S++ + E
Sbjct 298 KPTKGILLFGPPGTGKTMLAKAVATE--AGA-------------NFINISM-SSITSKGE 341
Query 293 KRMKEIISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILK---------VPLARGE 343
K +K + S A VI F+DE+ +++G + G +A + +K + E
Sbjct 342 KYVKAVFSLASKIAPSVI-FVDEVDSMLGRRENPGEHEAMRKMKNEFMINWDGLRTKDKE 400
Query 344 IVLVGATTLSEYKLYIEKDAAFCRRF-QKIVVEAPSKERTLGILQKVRTKYEDHHNMDIP 402
VLV A T + L D A RR ++++V P IL + K E ++D
Sbjct 401 RVLVLAATNRPFDL----DEAVIRRLPRRLMVNLPDSANRSKILSVILAKEEMAEDVD-- 454
Query 403 DEVLAAVVGLSDQY 416
L A+ ++D Y
Sbjct 455 ---LEAIANMTDGY 465
> At2g03670
Length=603
Score = 40.8 bits (94), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 59/208 (28%), Positives = 86/208 (41%), Gaps = 41/208 (19%)
Query 227 AQILSRKMTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSA 286
A+ L K + +L G PG GKT+++ + Q L+ LS S
Sbjct 48 ARTLGLKWPRGLLLYGPPGTGKTSLVRAVVQEC---------------DAHLIVLSPHSV 92
Query 287 MR---GEYEKRMKEIISYLQAHA---DEVILFIDEIHTLIGAGKAAGSMD---ASQILKV 337
R GE EK ++E + +HA ++FIDEI L A D ASQ+ +
Sbjct 93 HRAHAGESEKVLREAFAEASSHAVSDKPSVIFIDEIDVLCPRRDARREQDVRIASQLFTL 152
Query 338 -----PLARGEIVLVGATTLSEYKLYIEKDAAFCR--RFQKIV-VEAPSKERTLGILQKV 389
P + V+V A+T + D A R RF +V V P++E L ILQ
Sbjct 153 MDSNKPSSSAPRVVVVASTNRVDAI----DPALRRAGRFDALVEVSTPNEEDRLKILQ-- 206
Query 390 RTKYEDHHNMDIPDEVLAAVVGLSDQYI 417
Y N+D P L A+ + Y+
Sbjct 207 --LYTKKVNLD-PSVDLQAIAISCNGYV 231
> At2g27600
Length=435
Score = 40.4 bits (93), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 43/150 (28%), Positives = 63/150 (42%), Gaps = 25/150 (16%)
Query 232 RKMTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEY 291
R+ +A +L G PG GK+ + + +A + FS+ L S GE
Sbjct 162 RRPWRAFLLYGPPGTGKSYLAKAVATEA----------DSTFFSVSSSDLV--SKWMGES 209
Query 292 EKRMKEIISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPL--------ARGE 343
EK + + + A +I F+DEI +L G +AS+ +K L E
Sbjct 210 EKLVSNLFEMARESAPSII-FVDEIDSLCGTRGEGNESEASRRIKTELLVQMQGVGHNDE 268
Query 344 IVLVGATTLSEYKLYIEKDAAFCRRFQKIV 373
VLV A T + Y L D A RRF K +
Sbjct 269 KVLVLAATNTPYAL----DQAIRRRFDKRI 294
Score = 36.6 bits (83), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 26/93 (27%), Positives = 42/93 (45%), Gaps = 17/93 (18%)
Query 552 LTEEKKPIGAFLFLGSSGVGKTELAKALAEEVFHSEKNLIRLDMVEYQEAHSISRLIGPP 611
T +++P AFL G G GK+ LAKA+A E + ++ D+V
Sbjct 158 FTGKRRPWRAFLLYGPPGTGKSYLAKAVATEADSTFFSVSSSDLVS-------------- 203
Query 612 PGYMGNDEG--GQLTEAVRQKPHSVVLFDEVEN 642
+MG E L E R+ S++ DE+++
Sbjct 204 -KWMGESEKLVSNLFEMARESAPSIIFVDEIDS 235
> Hs11545833
Length=674
Score = 40.4 bits (93), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 39/128 (30%), Positives = 63/128 (49%), Gaps = 17/128 (13%)
Query 232 RKMTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEY 291
R K +L G PG GKT + + +A + GA FS+ SL+ S GE
Sbjct 431 RGPPKGILLFGPPGTGKTLIGKCIASQ--SGAT--------FFSISASSLT--SKWVGEG 478
Query 292 EKRMKEIISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPLARGEIVLVGATT 351
EK ++ + + + VI FIDEI +L+ + + G ++S+ +K + L GATT
Sbjct 479 EKMVRALFAVARCQQPAVI-FIDEIDSLL-SQRGDGEHESSRRIKTEFL---VQLDGATT 533
Query 352 LSEYKLYI 359
SE ++ +
Sbjct 534 SSEDRILV 541
> Hs18426902
Length=665
Score = 39.7 bits (91), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 41/152 (26%), Positives = 65/152 (42%), Gaps = 25/152 (16%)
Query 239 MLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYEKRMKEI 298
+L G PG GKT + +A N + S+ ++LSA +A + +K+
Sbjct 265 ILWGPPGCGKTTLAHIIAS------------NSKKHSIRFVTLSATNAKTNDVRDVIKQA 312
Query 299 ISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVP-LARGEIVLVGATTLSEYKL 357
+ + ILFIDEIH + Q +P + G I L+GATT +
Sbjct 313 QNEKSFFKRKTILFIDEIHRF---------NKSQQDTFLPHVECGTITLIGATTENP--- 360
Query 358 YIEKDAAFCRRFQKIVVEAPSKERTLGILQKV 389
+ +AA R + IV+E E + IL +
Sbjct 361 SFQVNAALLSRCRVIVLEKLPVEAMVTILMRA 392
> 7296674
Length=669
Score = 39.7 bits (91), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 42/169 (24%), Positives = 72/169 (42%), Gaps = 28/169 (16%)
Query 232 RKMTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEY 291
R+ + +++G PG GKT + + +A F++ +L+ S RGE
Sbjct 420 RRPWRGVLMVGPPGTGKTMLAKAVATEC----------GTTFFNVSSSTLT--SKYRGES 467
Query 292 EKRMKEIISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPL-----------A 340
EK ++ + + +A I FIDEI L + + +AS+ K L
Sbjct 468 EKLVRLLFEMARFYAPSTI-FIDEIDALCASRGSDSEHEASRRFKAELLIQMDGLNASMQ 526
Query 341 RGEIVLVGATTLSEYKLYIEKDAAFCRRFQKIVVEAPSKERTLGILQKV 389
++++V A T + + D AF RRF+K + E T L K+
Sbjct 527 EEKVIMVLAATNHPWDI----DEAFRRRFEKRIYIPLPNEGTRSALLKL 571
> CE19083
Length=412
Score = 39.7 bits (91), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 40/163 (24%), Positives = 67/163 (41%), Gaps = 16/163 (9%)
Query 527 LGKRVIGQDEAVQSVANAIRNHRAGLTEEKKPIGAFLFLGSSGVGKTELAKALAEEVFHS 586
++ GQ +SV ++I++H +KP+ F G +G GK + + + + S
Sbjct 128 FDNKIFGQHLMAESVVHSIKSHWHN-EHSQKPL-VLSFHGGTGTGKNYVTEIIVNNTYRS 185
Query 587 EKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDE---GGQLTEAVRQKPHSVVLFDEVENA 643
+ V Y A + P Y+ + + QL + R+ S+ +FDE +
Sbjct 186 G---MHSPFVNYFVATNN----FPNKKYIEDYKLELKDQLIRSARRCQRSIFIFDETDKL 238
Query 644 HKNLWSLLLPMLDEGHLTDTKNVRVDFTSTIIIMTSNIGQQYI 686
L ++ P LD VDF TI I SN G + I
Sbjct 239 QSELIQVIKPFLDYYPAV----FGVDFRKTIFIFLSNKGSKEI 277
> At1g80350
Length=523
Score = 39.7 bits (91), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 29/108 (26%), Positives = 52/108 (48%), Gaps = 13/108 (12%)
Query 232 RKMTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEY 291
R+ K ++ G PG GKT + + +A F++ +L+ S RGE
Sbjct 269 RRPWKGVLMFGPPGTGKTLLAKAVATEC----------GTTFFNVSSATLA--SKWRGES 316
Query 292 EKRMKEIISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPL 339
E+ ++ + +A+A I FIDEI +L + +G ++S+ +K L
Sbjct 317 ERMVRCLFDLARAYAPSTI-FIDEIDSLCNSRGGSGEHESSRRVKSEL 363
> At5g57710
Length=990
Score = 39.7 bits (91), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 27/119 (22%), Positives = 56/119 (47%), Gaps = 8/119 (6%)
Query 562 FLFLGSSGVGKTELAKALAEEVFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGG 621
LF G VGK ++ AL+ V+ + N I + + Q+A + + G
Sbjct 655 LLFSGPDRVGKRKMVSALSSLVYGT--NPIMIQLGSRQDAGDGNS------SFRGKTALD 706
Query 622 QLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDTKNVRVDFTSTIIIMTSN 680
++ E V++ P SV+L ++++ A + + +D G + D+ + + I +MT++
Sbjct 707 KIAETVKRSPFSVILLEDIDEADMLVRGSIKQAMDRGRIRDSHGREISLGNVIFVMTAS 765
> At4g24850
Length=442
Score = 39.7 bits (91), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 49/196 (25%), Positives = 84/196 (42%), Gaps = 28/196 (14%)
Query 233 KMTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYE 292
K K +L G PG GKT + + +A+ + S+ S S GE E
Sbjct 173 KPCKGILLFGPPGTGKTMLAKAVAKEADANFINISMS------------SITSKWFGEGE 220
Query 293 KRMKEIISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILK---------VPLARGE 343
K +K + S L + ++F+DE+ +++G + +AS+ +K + E
Sbjct 221 KYVKAVFS-LASKMSPSVIFVDEVDSMLGRREHPREHEASRKIKNEFMMHWDGLTTQERE 279
Query 344 IVLVGATTLSEYKLYIEKDAAFCRRF-QKIVVEAPSKERTLGILQKVRTKYEDHHNMDIP 402
VLV A T + L D A RR ++++V P IL+ + K + ++DI
Sbjct 280 RVLVLAATNRPFDL----DEAVIRRLPRRLMVGLPDTSNRAFILKVILAKEDLSPDLDI- 334
Query 403 DEVLAAVVGLSDQYIK 418
E+ + G S +K
Sbjct 335 GEIASMTNGYSGSDLK 350
Score = 33.5 bits (75), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 41/89 (46%), Gaps = 13/89 (14%)
Query 554 EEKKPIGAFLFLGSSGVGKTELAKALAEEVFHSEKNLIRLDMVEYQEAHSISRLIGPPPG 613
E KP L G G GKT LAKA+A+E ++ N I + M + S+ G
Sbjct 170 ELTKPCKGILLFGPPGTGKTMLAKAVAKE---ADANFINISM-----SSITSKWFGEGEK 221
Query 614 YMGNDEGGQLTEAVRQKPHSVVLFDEVEN 642
Y+ + A + P SV+ DEV++
Sbjct 222 YV----KAVFSLASKMSP-SVIFVDEVDS 245
> 7296781
Length=554
Score = 39.3 bits (90), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 43/168 (25%), Positives = 74/168 (44%), Gaps = 29/168 (17%)
Query 232 RKMTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEY 291
R+ K +++G PG GKT + + +A F++ +L+ S RGE
Sbjct 305 RRPWKGVLMVGPPGTGKTMLAKAVATEC----------GTTFFNVSSATLT--SKYRGES 352
Query 292 EKRMKEIISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPL-----------A 340
EK ++ + + +A I FIDEI +L + +AS+ +K L
Sbjct 353 EKMVRLLFEMARFYAPSTI-FIDEIDSLCSRRGSESEHEASRRVKSELLVQMDGVGGGEE 411
Query 341 RGEIVLVGATTLSEYKLYIEKDAAFCRRFQK-IVVEAPSKERTLGILQ 387
+ ++V+V A T + + D A RR +K I + PS E +L+
Sbjct 412 QAKVVMVLAATNFPWDI----DEALRRRLEKRIYIPLPSDEGREALLK 455
> Hs14150179
Length=939
Score = 38.9 bits (89), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 50/178 (28%), Positives = 74/178 (41%), Gaps = 22/178 (12%)
Query 239 MLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYEKRMKEI 298
+ G PG GKT V LA +G ++R+ S GE E++++
Sbjct 464 LFYGPPGTGKTLVARALANECSQG-------DKRVAFFMRKGADCLSKWVGESERQLR-- 514
Query 299 ISYLQAHA-DEVILFIDEIHTLIGAGKAAGSMDASQILKVPLA-------RGEIVLVGAT 350
+ + QA+ I+F DEI L + S I+ LA RGEIV++GAT
Sbjct 515 LLFDQAYQMRPSIIFFDEIDGLAPVRSSRQDQIHSSIVSTLLALMDGLDSRGEIVVIGAT 574
Query 351 T-LSEYKLYIEKDAAFCRRFQKIVVEAPSKERTLGILQKVRTKYEDHHNMDIPDEVLA 407
L + + F R F + P KE IL K+ T+ + +D E LA
Sbjct 575 NRLDSIDPALRRPGRFDREF---LFSLPDKEARKEIL-KIHTRDWNPKPLDTFLEELA 628
Score = 35.0 bits (79), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 31/115 (26%), Positives = 52/115 (45%), Gaps = 27/115 (23%)
Query 557 KPIGAFLFLGSSGVGKTELAKALAEEVFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYMG 616
+P LF G G GKT +A+ALA E +K R+ + A +S+ +G
Sbjct 458 QPPRGCLFYGPPGTGKTLVARALANECSQGDK---RVAFFMRKGADCLSKWVG------- 507
Query 617 NDEGGQLT----EAVRQKPHSVVLFDEV-----------ENAHKNLWSLLLPMLD 656
+ QL +A + +P S++ FDE+ + H ++ S LL ++D
Sbjct 508 -ESERQLRLLFDQAYQMRP-SIIFFDEIDGLAPVRSSRQDQIHSSIVSTLLALMD 560
> At1g05910
Length=1069
Score = 38.9 bits (89), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 47/181 (25%), Positives = 73/181 (40%), Gaps = 19/181 (10%)
Query 239 MLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYEKRMKEI 298
+L G PG GKT + LA + S R+ D+L S GE E+++K +
Sbjct 286 LLCGPPGTGKTLIARALACAASKAGQKVSFYMRK--GADVL-----SKWVGEAERQLKLL 338
Query 299 ISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPLA-------RGEIVLVGATT 351
Q + +I F DEI L + + I+ LA RG++VL+GAT
Sbjct 339 FEEAQRNQPSIIFF-DEIDGLAPVRSSKQEQIHNSIVSTLLALMDGLDSRGQVVLIGATN 397
Query 352 -LSEYKLYIEKDAAFCRRFQKIVVEAPSKERTLGILQKVRTKYEDHHNMDIPDEVLAAVV 410
+ + + F R F P E IL K++ ++ +E+ A V
Sbjct 398 RVDAIDGALRRPGRFDREFN---FSLPGCEARAEILDIHTRKWKHPPTRELKEELAATCV 454
Query 411 G 411
G
Sbjct 455 G 455
> SPBC56F2.07c
Length=809
Score = 38.9 bits (89), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 42/81 (51%), Gaps = 13/81 (16%)
Query 239 MLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYEKRMKEI 298
+L G PG GKT V+ +A N ++F++D S+ G + GE E R+++I
Sbjct 317 LLYGPPGTGKTMVMRAVAAEA----------NAQVFTIDGPSV-VGKYL-GETESRLRKI 364
Query 299 ISYLQAHADEVILFIDEIHTL 319
+AH +I FIDEI L
Sbjct 365 FEDARAHQPSII-FIDEIDAL 384
> At5g40090
Length=459
Score = 38.5 bits (88), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 29/100 (29%), Positives = 52/100 (52%), Gaps = 7/100 (7%)
Query 552 LTEEKKPIGAFLFLGSSGVGKTELAKALAEEVFHSEKNLIRLDMVEYQEAHSISRLIG-- 609
+ +E + IG + GS+GVGKT LA+ + E+F + + + LD VE + + + G
Sbjct 199 VNKEVRTIGIW---GSAGVGKTTLARYIYAEIFVNFQTHVFLDNVENMK-DKLLKFEGEE 254
Query 610 -PPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLW 648
P +G ++TEA R+ +++ D+V N + W
Sbjct 255 DPTVIISSYHDGHEITEARRKHRKILLIADDVNNMEQGKW 294
> Hs11875211
Length=616
Score = 38.1 bits (87), Expect = 0.072, Method: Compositional matrix adjust.
Identities = 47/156 (30%), Positives = 73/156 (46%), Gaps = 36/156 (23%)
Query 239 MLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYEKRMKEI 298
+L G PG GKT + + +A ES N F++ SL+ S GE EK +
Sbjct 379 LLFGPPGNGKTMLAKAVAA--------ES--NATFFNISAASLT--SKYVGEGEK----L 422
Query 299 ISYLQAHADEV---ILFIDEIHTLIGAGKAAGSMDASQILKVPL----------ARGEIV 345
+ L A A E+ I+FIDE+ +L+ + G DAS+ LK ++
Sbjct 423 VRALFAVARELQPSIIFIDEVDSLL-CERREGEHDASRRLKTEFLIEFDGVQSAGDDRVL 481
Query 346 LVGATTLSEYKLYIEKDAAFCRRF-QKIVVEAPSKE 380
++GAT + E D A RRF +++ V P++E
Sbjct 482 VMGATNRPQ-----ELDEAVLRRFIKRVYVSLPNEE 512
> CE20665
Length=1242
Score = 37.7 bits (86), Expect = 0.098, Method: Compositional matrix adjust.
Identities = 38/124 (30%), Positives = 55/124 (44%), Gaps = 17/124 (13%)
Query 236 KAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYEKRM 295
K + G PG GKT V LA GA + R+ D L S GE E+++
Sbjct 426 KGVVFYGPPGTGKTLVARALANECRRGANKVAFFMRK--GADCL-----SKWVGESERQL 478
Query 296 KEIISYLQAHA-DEVILFIDEIHTLIGAGKAAGSMDASQILKVPLA-------RGEIVLV 347
+ + + QA+A I+F DEI L + + I+ LA RGE+V++
Sbjct 479 R--LLFDQAYAMRPSIIFFDEIDGLAPVRSSKQDQIHASIVSTLLALMDGLDGRGEVVVI 536
Query 348 GATT 351
GAT
Sbjct 537 GATN 540
> YPL074w
Length=754
Score = 37.4 bits (85), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 48/184 (26%), Positives = 83/184 (45%), Gaps = 38/184 (20%)
Query 232 RKMTKAPMLLGEPGVGKTAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEY 291
R+ + +L G PG GKT + + +A ES N FS+ SL S GE
Sbjct 501 REPVRGMLLFGPPGTGKTMIAKAVAT--------ES--NSTFFSVSASSLL--SKYLGES 548
Query 292 EKRMKEIISYLQAHADEVILFIDEIHTLIGAGKAAGSMDASQILKVPL-----------A 340
EK ++ + Y+ I+FIDEI +++ A ++ ++S+ +K L A
Sbjct 549 EKLVRALF-YMAKKLSPSIIFIDEIDSMLTA-RSDNENESSRRIKTELLIQWSSLSSATA 606
Query 341 RGE---------IVLVGATTLSEYKLYIEKDAAFCRRFQKIVVEAPSKERTLGILQKVRT 391
+ E ++++GAT L + DAA R +K+ + P E L L+++
Sbjct 607 QSEDRNNTLDSRVLVLGATNLP----WAIDDAARRRFSRKLYIPLPDYETRLYHLKRLMA 662
Query 392 KYED 395
K ++
Sbjct 663 KQKN 666
> At4g24710
Length=400
Score = 37.4 bits (85), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 29/98 (29%), Positives = 48/98 (48%), Gaps = 15/98 (15%)
Query 239 MLLGEPGVGKTAVIEGLAQRIVEGAVPESLR-NRRIFSLDLLSLSAGSAMRGEYE----- 292
+L G PG GKT++ + LAQ++ S+R N R L+ ++A S +
Sbjct 158 LLHGPPGTGKTSLCKALAQKL-------SIRCNSRYPHCQLIEVNAHSLFSKWFSESGKL 210
Query 293 --KRMKEIISYLQAHADEVILFIDEIHTLIGAGKAAGS 328
K ++I ++ + V + IDE+ +L A KAA S
Sbjct 211 VAKLFQKIQEMVEEDGNLVFVLIDEVESLAAARKAALS 248
Lambda K H
0.317 0.135 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 21673112580
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40