bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0673_orf1
Length=243
Score E
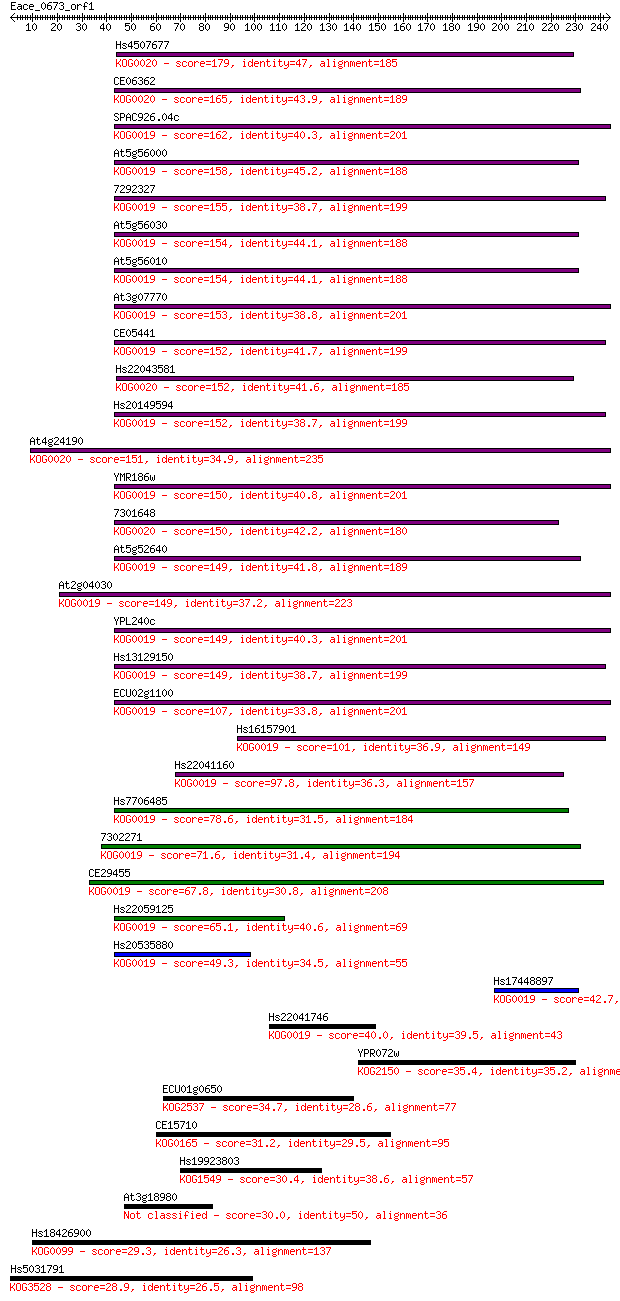
Sequences producing significant alignments: (Bits) Value
Hs4507677 179 4e-45
CE06362 165 6e-41
SPAC926.04c 162 6e-40
At5g56000 158 8e-39
7292327 155 7e-38
At5g56030 154 2e-37
At5g56010 154 2e-37
At3g07770 153 3e-37
CE05441 152 5e-37
Hs22043581 152 7e-37
Hs20149594 152 8e-37
At4g24190 151 1e-36
YMR186w 150 2e-36
7301648 150 3e-36
At5g52640 149 4e-36
At2g04030 149 6e-36
YPL240c 149 7e-36
Hs13129150 149 7e-36
ECU02g1100 107 3e-23
Hs16157901 101 1e-21
Hs22041160 97.8 2e-20
Hs7706485 78.6 1e-14
7302271 71.6 1e-12
CE29455 67.8 2e-11
Hs22059125 65.1 1e-10
Hs20535880 49.3 8e-06
Hs17448897 42.7 7e-04
Hs22041746 40.0 0.004
YPR072w 35.4 0.093
ECU01g0650 34.7 0.20
CE15710 31.2 1.9
Hs19923803 30.4 3.7
At3g18980 30.0 4.9
Hs18426900 29.3 7.0
Hs5031791 28.9 9.6
> Hs4507677
Length=803
Score = 179 bits (454), Expect = 4e-45, Method: Compositional matrix adjust.
Identities = 87/186 (46%), Positives = 126/186 (67%), Gaps = 1/186 (0%)
Query 44 DQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYYA 103
D F+KEFG N+ LG ED NR ++AK+L+F S+ PT+ SL+ YV RMKE Q IY+
Sbjct 482 DTFWKEFGTNIKLGVIEDHSNRTRLAKLLRFQSSHHPTDITSLDQYVERMKEKQDKIYFM 541
Query 104 AGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESIQKGNVTLEET 163
AG + ++ S P ++ KKGYEV++L E +DE CIQ L +FDGK+F+++ K V +E+
Sbjct 542 AGSSRKEAESSPFVERLLKKGYEVIYLTEPVDEYCIQALPEFDGKRFQNVAKEGVKFDES 601
Query 164 PDEKRQFKRLTKYYDPLLKWLK-KVLGPKVSKVEVSRRLVEAPCAVVASQWGYSAQMEKV 222
K + + K ++PLL W+K K L K+ K VS+RL E+PCA+VASQ+G+S ME++
Sbjct 602 EKTKESREAVEKEFEPLLNWMKDKALKDKIEKAVVSQRLTESPCALVASQYGWSGNMERI 661
Query 223 MKTQTF 228
MK Q +
Sbjct 662 MKAQAY 667
> CE06362
Length=760
Score = 165 bits (418), Expect = 6e-41, Method: Compositional matrix adjust.
Identities = 83/193 (43%), Positives = 126/193 (65%), Gaps = 4/193 (2%)
Query 43 YDQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYY 102
+D F+ EF N+ LG ED NR+++AK+L+F S+ + +L AYV RMKE Q AIYY
Sbjct 458 FDDFWSEFSTNIKLGVMEDPSNRMRLAKLLRFQSSNDADKTTTLAAYVERMKEKQDAIYY 517
Query 103 AAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESIQKGNVTLEE 162
AG + +++ + P ++ KGYEVLFL EA+DE CIQ + +++ KKF+++ K VT+++
Sbjct 518 MAGTSRKEVETSPFVERLIAKGYEVLFLTEAVDEYCIQAMPEYESKKFQNVAKEGVTIDD 577
Query 163 TPDEKRQFKRLTKYYDPLLKWLKKV-LGPKVSKVEVSRRLVEAPCAVVASQWGYSAQMEK 221
K K L + + PL WLK+ L + K VS+RLV++P A+VAS +G+S ME+
Sbjct 578 GEKAKEAHKGLEEEFKPLTDWLKETALKDLIEKAVVSQRLVKSPSALVASSYGWSGNMER 637
Query 222 VMKTQTFA---DP 231
+MK+Q +A DP
Sbjct 638 IMKSQAYAKAKDP 650
> SPAC926.04c
Length=704
Score = 162 bits (410), Expect = 6e-40, Method: Compositional matrix adjust.
Identities = 81/201 (40%), Positives = 123/201 (61%), Gaps = 0/201 (0%)
Query 43 YDQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYY 102
+ FY F +NL LG +ED NR +AK+L++ S SP + ISLE Y+++M E Q IY+
Sbjct 409 FKTFYDAFSKNLKLGIHEDAANRPALAKLLRYNSLNSPDDLISLEDYITKMPEHQKNIYF 468
Query 103 AAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESIQKGNVTLEE 162
GE+ Q + + P ++ F K ++VLF+++ +DE + +L +F+GKK +I K + LEE
Sbjct 469 ITGESKQAVENSPFLEIFRAKKFDVLFMVDPIDEYAVTQLKEFEGKKLVNITKDGLELEE 528
Query 163 TPDEKRQFKRLTKYYDPLLKWLKKVLGPKVSKVEVSRRLVEAPCAVVASQWGYSAQMEKV 222
T +EK ++L K Y+ K LK +LG KV KV VS ++V +PC + Q+G+SA ME++
Sbjct 529 TDEEKAAREKLEKEYEEFAKQLKTILGDKVEKVVVSNKIVGSPCLLTTGQYGWSANMERI 588
Query 223 MKTQTFADPMHLRLMRGHKVF 243
MK Q D M K F
Sbjct 589 MKAQALRDTSMSAYMSSRKTF 609
> At5g56000
Length=699
Score = 158 bits (400), Expect = 8e-39, Method: Compositional matrix adjust.
Identities = 85/188 (45%), Positives = 131/188 (69%), Gaps = 0/188 (0%)
Query 43 YDQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYY 102
Y++FY+ F +NL LG +ED +NR KIA++L+++S S E SL+ YV+RMKEGQ I+Y
Sbjct 405 YNKFYEAFSKNLKLGIHEDSQNRTKIAELLRYHSTKSGDELTSLKDYVTRMKEGQNEIFY 464
Query 103 AAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESIQKGNVTLEE 162
GE+ + + + P ++ KKGYEVL++++A+DE I +L +F+GKK S K + LEE
Sbjct 465 ITGESKKAVENSPFLEKLKKKGYEVLYMVDAIDEYAIGQLKEFEGKKLVSATKEGLKLEE 524
Query 163 TPDEKRQFKRLTKYYDPLLKWLKKVLGPKVSKVEVSRRLVEAPCAVVASQWGYSAQMEKV 222
T DEK++ + L + ++ L K +K VLG KV KV VS R+V++PC +V ++G++A ME++
Sbjct 525 TDDEKKKKEELKEKFEGLCKVIKDVLGDKVEKVIVSDRVVDSPCCLVTGEYGWTANMERI 584
Query 223 MKTQTFAD 230
MK Q D
Sbjct 585 MKAQALKD 592
> 7292327
Length=717
Score = 155 bits (392), Expect = 7e-38, Method: Compositional matrix adjust.
Identities = 77/199 (38%), Positives = 121/199 (60%), Gaps = 0/199 (0%)
Query 43 YDQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYY 102
Y +FY +F +NL LG +ED NR K+A L+F+++ S + SL YVSRMK+ Q +Y+
Sbjct 419 YKKFYDQFSKNLKLGVHEDSNNRAKLADFLRFHTSASGDDFCSLADYVSRMKDNQKHVYF 478
Query 103 AAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESIQKGNVTLEE 162
GE+ Q+ + ++ +G+EV+++ E +DE IQ L ++ GK+ S+ K + L E
Sbjct 479 ITGESKDQVSNSAFVERVKARGFEVVYMTEPIDEYVIQHLKEYKGKQLVSVTKEGLELPE 538
Query 163 TPDEKRQFKRLTKYYDPLLKWLKKVLGPKVSKVEVSRRLVEAPCAVVASQWGYSAQMEKV 222
EK++ + ++ L K +K +L KV KV VS RLV++PC +V SQ+G+SA ME++
Sbjct 539 DESEKKKREEDKAKFESLCKLMKSILDNKVEKVVVSNRLVDSPCCIVTSQFGWSANMERI 598
Query 223 MKTQTFADPMHLRLMRGHK 241
MK Q D + M G K
Sbjct 599 MKAQALRDTATMGYMAGKK 617
> At5g56030
Length=699
Score = 154 bits (388), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 83/188 (44%), Positives = 130/188 (69%), Gaps = 0/188 (0%)
Query 43 YDQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYY 102
Y++FY+ F +NL LG +ED +NR KIA++L+++S S E SL+ YV+RMKEGQ I+Y
Sbjct 405 YNKFYEAFSKNLKLGIHEDSQNRTKIAELLRYHSTKSGDELTSLKDYVTRMKEGQNDIFY 464
Query 103 AAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESIQKGNVTLEE 162
GE+ + + + P ++ KKG EVL++++A+DE I +L +F+GKK S K + L+E
Sbjct 465 ITGESKKAVENSPFLEKLKKKGIEVLYMVDAIDEYAIGQLKEFEGKKLVSATKEGLKLDE 524
Query 163 TPDEKRQFKRLTKYYDPLLKWLKKVLGPKVSKVEVSRRLVEAPCAVVASQWGYSAQMEKV 222
T DEK++ + L + ++ L K +K VLG KV KV VS R+V++PC +V ++G++A ME++
Sbjct 525 TEDEKKKKEELKEKFEGLCKVIKDVLGDKVEKVIVSDRVVDSPCCLVTGEYGWTANMERI 584
Query 223 MKTQTFAD 230
MK Q D
Sbjct 585 MKAQALRD 592
> At5g56010
Length=699
Score = 154 bits (388), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 83/188 (44%), Positives = 130/188 (69%), Gaps = 0/188 (0%)
Query 43 YDQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYY 102
Y++FY+ F +NL LG +ED +NR KIA++L+++S S E SL+ YV+RMKEGQ I+Y
Sbjct 405 YNKFYEAFSKNLKLGIHEDSQNRTKIAELLRYHSTKSGDELTSLKDYVTRMKEGQNDIFY 464
Query 103 AAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESIQKGNVTLEE 162
GE+ + + + P ++ KKG EVL++++A+DE I +L +F+GKK S K + L+E
Sbjct 465 ITGESKKAVENSPFLEKLKKKGIEVLYMVDAIDEYAIGQLKEFEGKKLVSATKEGLKLDE 524
Query 163 TPDEKRQFKRLTKYYDPLLKWLKKVLGPKVSKVEVSRRLVEAPCAVVASQWGYSAQMEKV 222
T DEK++ + L + ++ L K +K VLG KV KV VS R+V++PC +V ++G++A ME++
Sbjct 525 TEDEKKKKEELKEKFEGLCKVIKDVLGDKVEKVIVSDRVVDSPCCLVTGEYGWTANMERI 584
Query 223 MKTQTFAD 230
MK Q D
Sbjct 585 MKAQALRD 592
> At3g07770
Length=803
Score = 153 bits (387), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 78/203 (38%), Positives = 124/203 (61%), Gaps = 2/203 (0%)
Query 43 YDQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYY 102
Y++F+ FG++L LGC ED EN +IA +L+F+S+ S + ISL+ YV MK Q AIY+
Sbjct 500 YEKFWDNFGKHLKLGCIEDRENHKRIAPLLRFFSSQSENDMISLDEYVENMKPEQKAIYF 559
Query 103 AAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESIQKGNVTLEE 162
A ++ + P ++ +KG EVL+L+E +DE +Q L + K F I K ++ L
Sbjct 560 IASDSITSAKNAPFLEKMLEKGLEVLYLVEPIDEVAVQSLKAYKEKDFVDISKEDLDLGN 619
Query 163 TPDEKRQFKR--LTKYYDPLLKWLKKVLGPKVSKVEVSRRLVEAPCAVVASQWGYSAQME 220
+K + K + K + W+KK LG KV+ V++S RL +PC +V+ ++G+SA ME
Sbjct 620 MSGDKNEEKEAAVKKEFGQTCDWIKKRLGDKVASVQISNRLSSSPCVLVSGKFGWSANME 679
Query 221 KVMKTQTFADPMHLRLMRGHKVF 243
++MK Q+ D + L M+G +VF
Sbjct 680 RLMKAQSTGDTISLDYMKGRRVF 702
> CE05441
Length=702
Score = 152 bits (385), Expect = 5e-37, Method: Compositional matrix adjust.
Identities = 83/199 (41%), Positives = 127/199 (63%), Gaps = 1/199 (0%)
Query 43 YDQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYY 102
+ +FY++FG+NL LG +ED NR K++ L+ YS ++ EP SL+ YVSRMKE Q IYY
Sbjct 405 FKKFYEQFGKNLKLGIHEDSTNRKKLSDFLR-YSTSAGDEPTSLKEYVSRMKENQTQIYY 463
Query 103 AAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESIQKGNVTLEE 162
GE+ + + ++ +G+EVL++ + +DE C+Q+L ++DGKK S+ K + L E
Sbjct 464 ITGESKDVVAASAFVERVKSRGFEVLYMCDPIDEYCVQQLKEYDGKKLVSVTKEGLELPE 523
Query 163 TPDEKRQFKRLTKYYDPLLKWLKKVLGPKVSKVEVSRRLVEAPCAVVASQWGYSAQMEKV 222
T +EK++F+ Y+ L K +K +L KV KV VS RLV +PC +V S++G+SA ME++
Sbjct 524 TEEEKKKFEEDKVAYENLCKVIKDILEKKVEKVGVSNRLVSSPCCIVTSEYGWSANMERI 583
Query 223 MKTQTFADPMHLRLMRGHK 241
MK Q D + M K
Sbjct 584 MKAQALRDSSTMGYMAAKK 602
> Hs22043581
Length=329
Score = 152 bits (384), Expect = 7e-37, Method: Compositional matrix adjust.
Identities = 77/186 (41%), Positives = 116/186 (62%), Gaps = 1/186 (0%)
Query 44 DQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYYA 103
D F+KEFG N+ LG ED N+ +AK+L F S+ PT+ +L+ YV RMK Q IY+
Sbjct 113 DNFWKEFGTNIKLGVIEDHSNQTCLAKLLMFQSSHHPTDITTLDQYVERMKGKQDKIYFM 172
Query 104 AGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESIQKGNVTLEET 163
AG + ++ S P ++ KKG EV++L E +DE CIQ FDGK+F+++ K + +++
Sbjct 173 AGSSRKEAESPPSVERLLKKGCEVIYLTEPVDEHCIQAFPKFDGKRFQNVVKEGMKFDKS 232
Query 164 PDEKRQFKRLTKYYDPLLKWLK-KVLGPKVSKVEVSRRLVEAPCAVVASQWGYSAQMEKV 222
K + + K ++PLL W+K K L K+ K VS+ L E+ CA+VASQ+G S E++
Sbjct 233 EKTKESHEAVEKEFEPLLNWMKDKALKDKIEKAVVSQHLTESLCALVASQYGRSGNTERI 292
Query 223 MKTQTF 228
MK Q +
Sbjct 293 MKAQAY 298
> Hs20149594
Length=724
Score = 152 bits (383), Expect = 8e-37, Method: Compositional matrix adjust.
Identities = 77/199 (38%), Positives = 127/199 (63%), Gaps = 0/199 (0%)
Query 43 YDQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYY 102
Y +FY+ F +NL LG +ED NR +++++L+++++ S E SL YVSRMKE Q +IYY
Sbjct 426 YKKFYEAFSKNLKLGIHEDSTNRRRLSELLRYHTSQSGDEMTSLSEYVSRMKETQKSIYY 485
Query 103 AAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESIQKGNVTLEE 162
GE+ +Q+ + ++ K+G+EV+++ E +DE C+Q+L +FDGK S+ K + L E
Sbjct 486 ITGESKEQVANSAFVERVRKRGFEVVYMTEPIDEYCVQQLKEFDGKSLVSVTKEGLELPE 545
Query 163 TPDEKRQFKRLTKYYDPLLKWLKKVLGPKVSKVEVSRRLVEAPCAVVASQWGYSAQMEKV 222
+EK++ + ++ L K +K++L KV KV +S RLV +PC +V S +G++A ME++
Sbjct 546 DEEEKKKMEESKAKFENLCKLMKEILDKKVEKVTISNRLVSSPCCIVTSTYGWTANMERI 605
Query 223 MKTQTFADPMHLRLMRGHK 241
MK Q D + M K
Sbjct 606 MKAQALRDNSTMGYMMAKK 624
> At4g24190
Length=823
Score = 151 bits (381), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 82/238 (34%), Positives = 140/238 (58%), Gaps = 18/238 (7%)
Query 9 DANKKREEQQKQLEETSEEQEKEKIKKEMQQPSLYDQFYKEFGRNLMLGCYEDDENRLKI 68
D ++ ++++K +E++ E EK+ Y +F+ EFG+++ LG ED NR ++
Sbjct 486 DPDEIHDDEKKDVEKSGENDEKK---------GQYTKFWNEFGKSVKLGIIEDAANRNRL 536
Query 69 AKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYYAAGETAQQLLSQPQMQFFTKKGYEVL 128
AK+L+F + S + SL+ Y+ RMK+ Q I+Y G + +QL P ++ KKGYEV+
Sbjct 537 AKLLRFETTKSDGKLTSLDQYIKRMKKSQKDIFYITGSSKEQLEKSPFLERLIKKGYEVI 596
Query 129 FLLEAMDEPCIQRLGDFDGKKFESIQKGNVTL-EETPDE--KRQFKRLTKYYDPLLKWLK 185
F + +DE +Q L D++ KKF+++ K + + +++ D+ K FK LTK+ W
Sbjct 597 FFTDPVDEYLMQYLMDYEDKKFQNVSKEGLKVGKDSKDKELKEAFKELTKW------WKG 650
Query 186 KVLGPKVSKVEVSRRLVEAPCAVVASQWGYSAQMEKVMKTQTFADPMHLRLMRGHKVF 243
+ V V++S RL + PC VV S++G+SA ME++M++QT +D MRG +V
Sbjct 651 NLASENVDDVKISNRLADTPCVVVTSKFGWSANMERIMQSQTLSDANKQAYMRGKRVL 708
> YMR186w
Length=705
Score = 150 bits (380), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 82/201 (40%), Positives = 119/201 (59%), Gaps = 1/201 (0%)
Query 43 YDQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYY 102
+D+FY F +N+ LG +ED +NR +AK+L++ S S E SL YV+RM E Q IYY
Sbjct 410 FDKFYSAFAKNIKLGVHEDTQNRAALAKLLRYNSTKSVDELTSLTDYVTRMPEHQKNIYY 469
Query 103 AAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESIQKGNVTLEE 162
GE+ + + P + K +EVLFL + +DE +L +F+GK I K + LEE
Sbjct 470 ITGESLKAVEKSPFLDALKAKNFEVLFLTDPIDEYAFTQLKEFEGKTLVDITK-DFELEE 528
Query 163 TPDEKRQFKRLTKYYDPLLKWLKKVLGPKVSKVEVSRRLVEAPCAVVASQWGYSAQMEKV 222
T +EK + ++ K Y+PL K LK +LG +V KV VS +L++AP A+ Q+G+SA ME++
Sbjct 529 TDEEKAEREKEIKEYEPLTKALKDILGDQVEKVVVSYKLLDAPAAIRTGQFGWSANMERI 588
Query 223 MKTQTFADPMHLRLMRGHKVF 243
MK Q D M K F
Sbjct 589 MKAQALRDSSMSSYMSSKKTF 609
> 7301648
Length=787
Score = 150 bits (378), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 76/181 (41%), Positives = 113/181 (62%), Gaps = 2/181 (1%)
Query 43 YDQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYY 102
Y++F+KEF N+ LG ED NR ++AK+L+F ++ SL Y RMK Q IYY
Sbjct 479 YEKFWKEFSTNIKLGVMEDPSNRSRLAKLLRFQTSNGKG-VTSLAEYKERMKAKQEHIYY 537
Query 103 AAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESIQKGNVTLEE 162
AG ++ P ++ KGYEVL+L+EA+DE CI L +FDGKKF+++ K L E
Sbjct 538 IAGANRAEVEKSPFVERLLSKGYEVLYLVEAVDEYCISALPEFDGKKFQNVAKEGFQLNE 597
Query 163 TPDEKRQFKRLTKYYDPLLKWLKKV-LGPKVSKVEVSRRLVEAPCAVVASQWGYSAQMEK 221
+ K+ F+ L ++PL+KWL V L ++SK +VS RL +PCA+VA +G++ ME+
Sbjct 598 SEKSKKNFESLKSTFEPLVKWLNDVALKDQISKAQVSERLSNSPCALVAGVFGWTGNMER 657
Query 222 V 222
+
Sbjct 658 L 658
> At5g52640
Length=705
Score = 149 bits (377), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 79/190 (41%), Positives = 127/190 (66%), Gaps = 1/190 (0%)
Query 43 YDQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYY 102
Y +FY+ F +NL LG +ED +NR KIA +L+++S S E S + YV+RMKEGQ I+Y
Sbjct 411 YTKFYEAFSKNLKLGIHEDSQNRGKIADLLRYHSTKSGDEMTSFKDYVTRMKEGQKDIFY 470
Query 103 AAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESIQK-GNVTLE 161
GE+ + + + P ++ K+GYEVL++++A+DE + +L ++DGKK S K G +
Sbjct 471 ITGESKKAVENSPFLERLKKRGYEVLYMVDAIDEYAVGQLKEYDGKKLVSATKEGLKLED 530
Query 162 ETPDEKRQFKRLTKYYDPLLKWLKKVLGPKVSKVEVSRRLVEAPCAVVASQWGYSAQMEK 221
ET +EK++ + K ++ L K +K++LG KV KV VS R+V++PC +V ++G++A ME+
Sbjct 531 ETEEEKKKREEKKKSFENLCKTIKEILGDKVEKVVVSDRIVDSPCCLVTGEYGWTANMER 590
Query 222 VMKTQTFADP 231
+MK Q D
Sbjct 591 IMKAQALRDS 600
> At2g04030
Length=780
Score = 149 bits (375), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 83/223 (37%), Positives = 128/223 (57%), Gaps = 12/223 (5%)
Query 21 LEETSEEQEKEKIKKEMQQPSLYDQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSP 80
++E SE + KE KK F++ FGR L LGC ED N +I +L+F+S+ +
Sbjct 465 IQEISESENKEDYKK----------FWENFGRFLKLGCIEDTGNHKRITPLLRFFSSKNE 514
Query 81 TEPISLEAYVSRMKEGQPAIYYAAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQ 140
E SL+ Y+ M E Q AIYY A ++ + S P ++ +K EVL+L+E +DE IQ
Sbjct 515 EELTSLDDYIENMGENQKAIYYLATDSLKSAKSAPFLEKLIQKDIEVLYLVEPIDEVAIQ 574
Query 141 RLGDFDGKKFESIQKGNVTLEETPDEKRQFKRLTKYYDPLLKWLKKVLGPKVSKVEVSRR 200
L + KKF I K ++ L + + K + + ++ L W+K+ LG KV+KV+VS R
Sbjct 575 NLQTYKEKKFVDISKEDLELGDEDEVKD--REAKQEFNLLCDWIKQQLGDKVAKVQVSNR 632
Query 201 LVEAPCAVVASQWGYSAQMEKVMKTQTFADPMHLRLMRGHKVF 243
L +PC +V+ ++G+SA ME++MK Q D L MRG ++
Sbjct 633 LSSSPCVLVSGKFGWSANMERLMKAQALGDTSSLEFMRGRRIL 675
> YPL240c
Length=709
Score = 149 bits (375), Expect = 7e-36, Method: Compositional matrix adjust.
Identities = 81/201 (40%), Positives = 120/201 (59%), Gaps = 1/201 (0%)
Query 43 YDQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYY 102
+++FY F +N+ LG +ED +NR +AK+L++ S S E SL YV+RM E Q IYY
Sbjct 414 FEKFYSAFSKNIKLGVHEDTQNRAALAKLLRYNSTKSVDELTSLTDYVTRMPEHQKNIYY 473
Query 103 AAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESIQKGNVTLEE 162
GE+ + + P + K +EVLFL + +DE +L +F+GK I K + LEE
Sbjct 474 ITGESLKAVEKSPFLDALKAKNFEVLFLTDPIDEYAFTQLKEFEGKTLVDITK-DFELEE 532
Query 163 TPDEKRQFKRLTKYYDPLLKWLKKVLGPKVSKVEVSRRLVEAPCAVVASQWGYSAQMEKV 222
T +EK + ++ K Y+PL K LK++LG +V KV VS +L++AP A+ Q+G+SA ME++
Sbjct 533 TDEEKAEREKEIKEYEPLTKALKEILGDQVEKVVVSYKLLDAPAAIRTGQFGWSANMERI 592
Query 223 MKTQTFADPMHLRLMRGHKVF 243
MK Q D M K F
Sbjct 593 MKAQALRDSSMSSYMSSKKTF 613
> Hs13129150
Length=732
Score = 149 bits (375), Expect = 7e-36, Method: Compositional matrix adjust.
Identities = 77/199 (38%), Positives = 126/199 (63%), Gaps = 0/199 (0%)
Query 43 YDQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYY 102
Y +FY++F +N+ LG +ED +NR K++++L++Y++ S E +SL+ Y +RMKE Q IYY
Sbjct 434 YKKFYEQFSKNIKLGIHEDSQNRKKLSELLRYYTSASGDEMVSLKDYCTRMKENQKHIYY 493
Query 103 AAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESIQKGNVTLEE 162
GET Q+ + ++ K G EV++++E +DE C+Q+L +F+GK S+ K + L E
Sbjct 494 ITGETKDQVANSAFVERLRKHGLEVIYMIEPIDEYCVQQLKEFEGKTLVSVTKEGLELPE 553
Query 163 TPDEKRQFKRLTKYYDPLLKWLKKVLGPKVSKVEVSRRLVEAPCAVVASQWGYSAQMEKV 222
+EK++ + ++ L K +K +L KV KV VS RLV +PC +V S +G++A ME++
Sbjct 554 DEEEKKKQEEKKTKFENLCKIMKDILEKKVEKVVVSNRLVTSPCCIVTSTYGWTANMERI 613
Query 223 MKTQTFADPMHLRLMRGHK 241
MK Q D + M K
Sbjct 614 MKAQALRDNSTMGYMAAKK 632
> ECU02g1100
Length=690
Score = 107 bits (266), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 68/204 (33%), Positives = 104/204 (50%), Gaps = 8/204 (3%)
Query 43 YDQFYKEFGRNLMLGCYEDDENRLK-IAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIY 101
Y FYKEFG L + E E + AK L++++ S E ISL+ YV RM Q IY
Sbjct 420 YKTFYKEFGNCLKMAIGEASEGQQDGYAKCLRYFTTKSGEEAISLDTYVERMAPNQKQIY 479
Query 102 YAAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESIQKGNVTLE 161
G + +Q+ S P + F K YEV+++ E MDE ++ L + G + I V
Sbjct 480 VITGLSKEQVKSNPALDAFQK--YEVIYMHEVMDEVMLRGLKKYKGHTIQRITSEGV--- 534
Query 162 ETPDEKRQFKRLTKYYDPLLKWLKKVLGPKVSKVEVSRRLVEAPCAVVASQWGYSAQMEK 221
E P+++ + + K ++ K +K +L KV KV V+ RLV P + +++ S ME
Sbjct 535 ELPEDEASNEEVVKSFEEFCKKVKDILSSKVEKVTVNPRLVSVPAVISTTKYSLSGTMEN 594
Query 222 VMKTQ--TFADPMHLRLMRGHKVF 243
+MK+Q T A+P K+F
Sbjct 595 IMKSQPVTEANPFAAMTAVSKKIF 618
> Hs16157901
Length=249
Score = 101 bits (252), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 55/149 (36%), Positives = 92/149 (61%), Gaps = 0/149 (0%)
Query 93 MKEGQPAIYYAAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFES 152
MKE Q + YY GE+ +Q+ + ++ K+G+EV+++ E +DE C+Q+L +FDGK S
Sbjct 1 MKETQKSTYYITGESKEQVANSAFVERVRKQGFEVVYMTEPIDEYCVQQLKEFDGKSLVS 60
Query 153 IQKGNVTLEETPDEKRQFKRLTKYYDPLLKWLKKVLGPKVSKVEVSRRLVEAPCAVVASQ 212
+ K + L E +EK++ + + ++ L K +K++L KV KV +S RLV +PC +V S
Sbjct 61 VTKEGLELPEDEEEKKKMEESKEKFENLCKLMKEILDKKVEKVTISNRLVSSPCCIVTST 120
Query 213 WGYSAQMEKVMKTQTFADPMHLRLMRGHK 241
+G++A ME++MK Q D + M K
Sbjct 121 YGWTANMEQIMKAQALRDNSTMGYMMAKK 149
> Hs22041160
Length=343
Score = 97.8 bits (242), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 57/157 (36%), Positives = 90/157 (57%), Gaps = 3/157 (1%)
Query 68 IAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYYAAGETAQQLLSQPQMQFFTKKGYEV 127
I + L F S E ISL YVSRMKE Q +IYY GE+ +Q+ + ++ K+ V
Sbjct 189 IPECLNFIRGVSGDEMISLLEYVSRMKEIQKSIYYITGESKEQVANSAFVEQVWKRDSRV 248
Query 128 LFLLEAMDEPCIQRLGDFDGKKFESIQKGNVTLEETPDEKRQFKRLTKYYDPLLKWLKKV 187
+++ E +D +L +FDGK S+ K + L E +EK++ + ++ L K++K+
Sbjct 249 VYMTEPIDG---YQLKEFDGKSLVSVTKEGLELPEDGEEKKRMEERKAKFENLCKFMKET 305
Query 188 LGPKVSKVEVSRRLVEAPCAVVASQWGYSAQMEKVMK 224
L KV V VS RLV + C +V S + ++A ME++MK
Sbjct 306 LDKKVEMVTVSNRLVSSSCCIVTSTYSWTANMEQIMK 342
> Hs7706485
Length=704
Score = 78.6 bits (192), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 58/196 (29%), Positives = 98/196 (50%), Gaps = 13/196 (6%)
Query 43 YDQFYKEFGRNLMLGCYEDDENRLK--IAKVLKFYSNTSPTEPI-SLEAYVSRMKEGQPA 99
Y +F++++G + G E +K IAK+L++ S+ P+ + SL Y SRM+ G
Sbjct 437 YAKFFEDYGLFMREGIVTATEQEVKEDIAKLLRYESSALPSGQLTSLSEYASRMRAGTRN 496
Query 100 IYYAAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESIQKGNVT 159
IYY P + KK EVLF E DE + L +FD KK S++ ++
Sbjct 497 IYYLCAPNRHLAEHSPYYEAMKKKDTEVLFCFEQFDELTLLHLREFDKKKLISVET-DIV 555
Query 160 LEETPDEKRQFKRLT------KYYDPLLKWLKKVLGPKVSKVEVSRRLVEAPCAVVASQW 213
++ +EK + + K + L+ W++ VLG +V+ V+V+ RL P V +
Sbjct 556 VDHYKEEKFEDRSPAAECLSEKETEELMAWMRNVLGSRVTNVKVTLRLDTHPAMVTVLEM 615
Query 214 GYS---AQMEKVMKTQ 226
G + +M+++ KTQ
Sbjct 616 GAARHFLRMQQLAKTQ 631
> 7302271
Length=691
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 61/207 (29%), Positives = 95/207 (45%), Gaps = 22/207 (10%)
Query 38 QQPSLYDQFYKEFGRNLMLGCY--EDDENRLKIAKVLKFYSNTSPTEP--ISLEAYVSRM 93
+QP Y+ FY+++G L G D + +IAK+L+F S+ S T +SLE Y + +
Sbjct 420 KQPEEYEAFYRDYGLFLKEGIVTSSDASEKEEIAKLLRFESSKSETASGRMSLEEYYNAV 479
Query 94 KEGQPAIYYAAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGKKFESI 153
Q IYY A S P + K+ VLF E DE + +LG F KK S+
Sbjct 480 PAEQQNIYYLAAPNRVLAESSPYYESLKKRNELVLFCYEPYDELVLMQLGKFKSKKLVSV 539
Query 154 QKGNVTLEETPDEKRQFKRLTKY---------YDPLLKWLKKVLGPKVSKVEVSRRLVEA 204
+K E +E ++ T + D L+ WL++ L +V KV+ + RL
Sbjct 540 EK------EMREESKETTATTDFGEGSLLRSELDTLIPWLEEELRGQVIKVKATTRLDTH 593
Query 205 PCAVVASQWGYSAQMEKVMKTQTFADP 231
PC + + A ++TQ+ P
Sbjct 594 PCVITVEEM---AAARHFIRTQSHQVP 617
> CE29455
Length=657
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 64/218 (29%), Positives = 97/218 (44%), Gaps = 15/218 (6%)
Query 33 IKKEMQQ-PSLYDQFYKEFGRNLMLGCYEDDENRLK--IAKVLKFYSNTSPT-EPISLEA 88
++ EM++ P Y +F+K + G + + +K +AK+L F S++ E SL
Sbjct 374 LQSEMKKDPVKYSEFFKNYSLYFKEGVVTEQDQGVKEDVAKLLLFESSSKKAGELTSLGD 433
Query 89 YVSRMKEGQPAIYYAAGETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGK 148
YV RM+EGQ IYY Q S P + + EVLFL + DE LG F K
Sbjct 434 YVKRMQEGQKEIYYMYANNRQLAESSPYYEVIKSQNKEVLFLYDPADEVVFLGLGQFGMK 493
Query 149 KFESIQKGNVTLEETPDEKRQFKRLTKYY-----DPLLKWLKKVLGP-KVSKVEVSRRLV 202
+ ++K EE K+ TK + LL W+K+ LG +VS++ + R
Sbjct 494 QLVPVEKW--AQEEAEKTGTDDKKDTKDFRDNEKKELLDWMKETLGSVRVSEISGNHRPS 551
Query 203 EAPCAVVASQWGYSAQMEKVMKTQTFADPMHLRLMRGH 240
E P V G + ++T D HL + H
Sbjct 552 EHPVMVTVLDMGAARHF---LRTGEIKDMEHLVYFKPH 586
> Hs22059125
Length=192
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 28/69 (40%), Positives = 47/69 (68%), Gaps = 0/69 (0%)
Query 43 YDQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYY 102
Y +FY+ F +NL LG +ED N+ +++++L ++++ S E SL Y+S MKE Q +IYY
Sbjct 60 YKKFYEAFSKNLNLGIHEDSTNQGRLSELLCYHTSQSEDEMTSLSEYLSHMKEAQKSIYY 119
Query 103 AAGETAQQL 111
GE+ +Q+
Sbjct 120 ITGESKEQV 128
> Hs20535880
Length=340
Score = 49.3 bits (116), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 41/55 (74%), Gaps = 0/55 (0%)
Query 43 YDQFYKEFGRNLMLGCYEDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQ 97
Y +FY++F N+ LG +ED +N+ K++++L++Y++ S E +SL+ Y +R+++ +
Sbjct 283 YKKFYEQFSENIKLGIHEDSQNQKKLSELLRYYTSASGDEMVSLKDYCTRIRKTR 337
> Hs17448897
Length=101
Score = 42.7 bits (99), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 23/34 (67%), Gaps = 0/34 (0%)
Query 197 VSRRLVEAPCAVVASQWGYSAQMEKVMKTQTFAD 230
+ RLV +PC +V S +G++A ME++MK Q D
Sbjct 3 IFNRLVSSPCCIVTSTYGWTANMEQIMKAQALRD 36
> Hs22041746
Length=1595
Score = 40.0 bits (92), Expect = 0.004, Method: Composition-based stats.
Identities = 17/43 (39%), Positives = 28/43 (65%), Gaps = 0/43 (0%)
Query 106 ETAQQLLSQPQMQFFTKKGYEVLFLLEAMDEPCIQRLGDFDGK 148
ET Q+ + +Q K G EV++ +E +DE C+Q+L +F+GK
Sbjct 855 ETKDQVANSTIVQRLWKHGLEVIYTIEPIDEYCVQQLKEFEGK 897
> YPR072w
Length=560
Score = 35.4 bits (80), Expect = 0.093, Method: Compositional matrix adjust.
Identities = 31/93 (33%), Positives = 48/93 (51%), Gaps = 12/93 (12%)
Query 142 LGDFDG--KKFESIQKGNVTLEET--PDEKRQFKRLTKYYDPLLKWL-KKVLGPKVSKVE 196
+ DFD +KF+S N + E D KR+ K+L K+ D + WL K+ + K S +
Sbjct 21 IEDFDDIYEKFQSTDPSNSSHREKLESDLKREIKKLQKHRDQIKTWLSKEDVKDKQSVLM 80
Query 197 VSRRLVEAPCAVVASQWGYSAQMEKVMKTQTFA 229
+RRL+E S +EK+MKT+ F+
Sbjct 81 TNRRLIENGMERFKS-------VEKLMKTKQFS 106
> ECU01g0650
Length=530
Score = 34.7 bits (78), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 22/92 (23%), Positives = 41/92 (44%), Gaps = 15/92 (16%)
Query 63 ENRLKIAKVLKFYSNTSPTEPI------------SLEAYVSRMKEGQPAIYYAAGETAQQ 110
E+RL I VL +YSN + + + ++ +VS +E IY+
Sbjct 317 ESRLSIGVVLSYYSNNAAVDVLPPESFKVVMAQTGVKNFVSAAREFDVGIYFEPNGHGSV 376
Query 111 LLSQP---QMQFFTKKGYEVLFLLEAMDEPCI 139
SQ +++ + K + +L +L + +PCI
Sbjct 377 CFSQACIDEIEKGSTKSHAILKILANLFDPCI 408
> CE15710
Length=1186
Score = 31.2 bits (69), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 42/99 (42%), Gaps = 12/99 (12%)
Query 60 EDDENRLKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYYAAGETAQQLLSQPQMQF 119
EDDEN+ K + SP I LE S Q AI+ ET +++
Sbjct 712 EDDENQDKDTVAPSAENLDSPPSDIPLETLSSIPSASQSAIFLQDSETGKEM-------- 763
Query 120 FTKKGYEV-LFLLEAMDEPCI---QRLGDFDGKKFESIQ 154
K +V + +LEA D P +DG+K E+++
Sbjct 764 HVPKAEDVGVVVLEASDSPVALEGNNEASYDGQKIENLE 802
> Hs19923803
Length=445
Score = 30.4 bits (67), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 33/71 (46%), Gaps = 14/71 (19%)
Query 70 KVLKFYSNTSPTEPISLEAYVSRMKE--GQPAIYYAAGETAQQLLS------------QP 115
KV Y+ T+P EP ++A M E G P+ Y+AG A+ +++ +P
Sbjct 31 KVYMDYNATTPLEPEVIQAMTKAMWEAWGNPSSPYSAGRKAKDIINAARESLAKMIGGKP 90
Query 116 QMQFFTKKGYE 126
Q FT G E
Sbjct 91 QDIIFTSGGTE 101
> At3g18980
Length=415
Score = 30.0 bits (66), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 23/40 (57%), Gaps = 4/40 (10%)
Query 47 YKEFGRNLMLG-CYEDDENRL---KIAKVLKFYSNTSPTE 82
+KE+ +LG CY+ DEN KI K+L FY N TE
Sbjct 143 WKEYEVRFVLGYCYQQDENNSCSKKIYKILCFYPNGQDTE 182
> Hs18426900
Length=380
Score = 29.3 bits (64), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 36/142 (25%), Positives = 64/142 (45%), Gaps = 24/142 (16%)
Query 10 ANKKREEQQKQLEETSEEQEKEKIKKEMQQPSLYDQFYKEFGRNLMLGCYEDDEN----R 65
N K E+Q+ EE ++ + +KI+K++Q+ Q Y+ R L+LG E ++ +
Sbjct 5 GNSKTEDQRN--EEKAQREANKKIEKQLQKDK---QVYRATHRLLLLGAGESGKSTIVKQ 59
Query 66 LKIAKVLKFYSNTSPTEPISLEAYVSRMKEGQPAIYYAAGETAQQL-LSQPQMQFFTKKG 124
++I V F N + ++ + +KE I A + L+ P+ QF
Sbjct 60 MRILHVNGF--NGDSEKATKVQDIKNNLKEAIETIVAAMSNLVPPVELANPENQF----- 112
Query 125 YEVLFLLEAMDEPCIQRLGDFD 146
V ++L M+ P DFD
Sbjct 113 -RVDYILSVMNVP------DFD 127
> Hs5031791
Length=1582
Score = 28.9 bits (63), Expect = 9.6, Method: Composition-based stats.
Identities = 26/99 (26%), Positives = 42/99 (42%), Gaps = 24/99 (24%)
Query 1 ETVRKMFLDANKKREEQQKQLEETSEEQEKEKIKKEMQQPSLYDQFYKEFGRNLMLGCYE 60
E V + FLD K QQK++E++ E E E P L +
Sbjct 824 ELVDEPFLDLGKSFHSQQKEIEQSKEAWEMH----EFLTPRLQEM--------------- 864
Query 61 DDENRLKIAKVLKFYSNTSPTEPISLEAY-VSRMKEGQP 98
D+E + + + + Y + SP S+E Y +S ++E P
Sbjct 865 DEEREMLVDEEYELYQDPSP----SMELYPLSHIQEATP 899
Lambda K H
0.316 0.132 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4867742342
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40