bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0670_orf1
Length=147
Score E
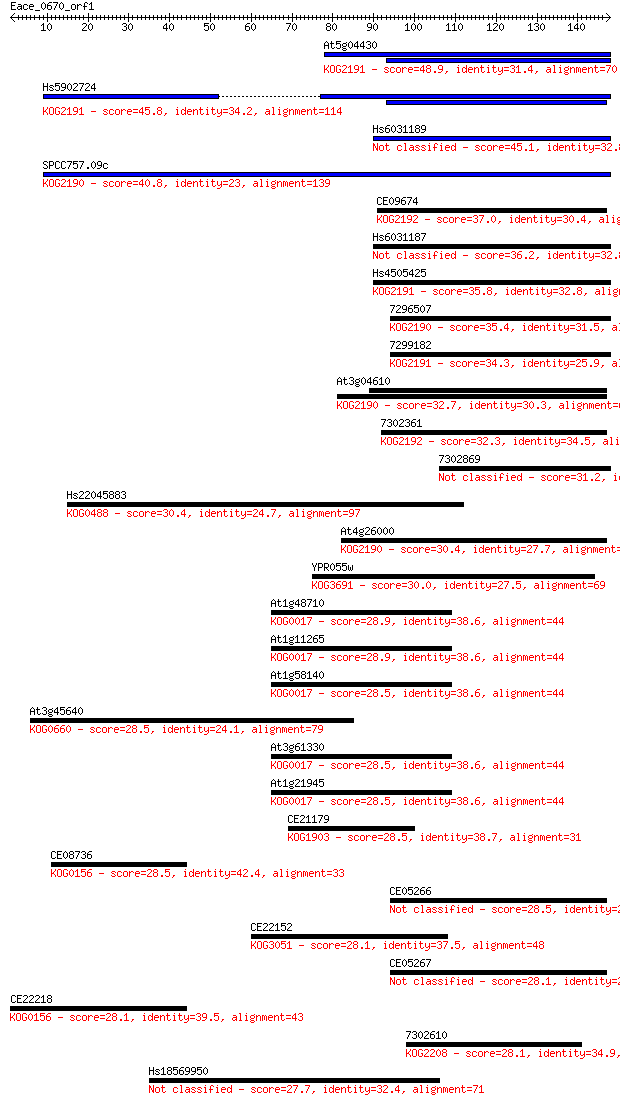
Sequences producing significant alignments: (Bits) Value
At5g04430 48.9 3e-06
Hs5902724 45.8 3e-05
Hs6031189 45.1 6e-05
SPCC757.09c 40.8 0.001
CE09674 37.0 0.013
Hs6031187 36.2 0.024
Hs4505425 35.8 0.031
7296507 35.4 0.039
7299182 34.3 0.097
At3g04610 32.7 0.25
7302361 32.3 0.34
7302869 31.2 0.66
Hs22045883 30.4 1.1
At4g26000 30.4 1.4
YPR055w 30.0 1.8
At1g48710 28.9 3.9
At1g11265 28.9 4.1
At1g58140 28.5 4.3
At3g45640 28.5 4.5
At3g61330 28.5 4.7
At1g21945 28.5 4.7
CE21179 28.5 5.0
CE08736 28.5 5.1
CE05266 28.5 5.3
CE22152 28.1 5.5
CE05267 28.1 5.9
CE22218 28.1 6.4
7302610 28.1 6.4
Hs18569950 27.7 9.2
> At5g04430
Length=313
Score = 48.9 bits (115), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 39/70 (55%), Gaps = 0/70 (0%)
Query 78 EPLEDERPVSLPGPFHMKLLIDPALANSLIGENSQTITKVAEECGCSMHVLSSKNKFPDS 137
EP + S P H++ L+ A A S+IG+ TIT+ + G + + ++ FP +
Sbjct 21 EPHDSSEADSAEKPTHIRFLVSNAAAGSVIGKGGSTITEFQAKSGARIQLSRNQEFFPGT 80
Query 138 QDRLLLMSGS 147
DR++++SGS
Sbjct 81 TDRIIMISGS 90
Score = 28.5 bits (62), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 14/55 (25%), Positives = 26/55 (47%), Gaps = 0/55 (0%)
Query 93 HMKLLIDPALANSLIGENSQTITKVAEECGCSMHVLSSKNKFPDSQDRLLLMSGS 147
++L++ + +IG+ TI EE + + N F DRL+ +SG+
Sbjct 122 RIRLVVPNSSCGGIIGKGGATIKSFIEESKAGIKISPLDNTFYGLSDRLVTLSGT 176
> Hs5902724
Length=492
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 44/85 (51%), Gaps = 14/85 (16%)
Query 77 EEPLEDERPVSLP--------------GPFHMKLLIDPALANSLIGENSQTITKVAEECG 122
E P +RP+ P G + +K+LI A S+IG+ QTI ++ +E G
Sbjct 4 EAPDSRKRPLETPPEVVCTKRSNTGEEGEYFLKVLIPSYAAGSIIGKGGQTIVQLQKETG 63
Query 123 CSMHVLSSKNKFPDSQDRLLLMSGS 147
++ + SK+ +P + +R+ L+ G+
Sbjct 64 ATIKLSKSKDFYPGTTERVCLVQGT 88
Score = 32.3 bits (72), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 9 NPHERIISVLGPLEQVTIAIKAMLPYIHADPNQSQSVHQSYGH 51
N ER+++V G EQV A+ A++ + DP S ++ SY +
Sbjct 174 NLQERVVTVSGEPEQVHKAVSAIVQKVQEDPQSSSCLNISYAN 216
Score = 27.7 bits (60), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 30/54 (55%), Gaps = 1/54 (1%)
Query 93 HMKLLIDPALANSLIGENSQTITKVAEECGCSMHVLSSKNKFPDSQDRLLLMSG 146
KL++ + A +IG+ T+ V E+ G + LS K + + Q+R++ +SG
Sbjct 132 QAKLIVPNSTAGLIIGKGGATVKAVMEQSGAWVQ-LSQKPEGINLQERVVTVSG 184
> Hs6031189
Length=181
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 37/58 (63%), Gaps = 0/58 (0%)
Query 90 GPFHMKLLIDPALANSLIGENSQTITKVAEECGCSMHVLSSKNKFPDSQDRLLLMSGS 147
G + +K+LI A S+IG+ QTI ++ +E G ++ + SK+ +P + +R+ L+ G+
Sbjct 48 GQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKDFYPGTTERVCLIQGT 105
> SPCC757.09c
Length=398
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 32/139 (23%), Positives = 56/139 (40%), Gaps = 41/139 (29%)
Query 9 NPHERIISVLGPLEQVTIAIKAMLPYIHADPNQSQSVHQSYGHGYKLKMPSWAETGVLQE 68
N H+R++++ GPLE V A Y+ + +A+ +
Sbjct 135 NVHDRVLTISGPLENVVRA-------------------------YRFIIDIFAKNSTNPD 169
Query 69 GMNMEALKEEPLEDERPVSLPGPFHMKLLIDPALANSLIGENSQTITKVAEECGCSMHVL 128
G P P ++LLI +L S+IG N I + ++C C M +
Sbjct 170 GT--------------PSDANTPRKLRLLIAHSLMGSIIGRNGLRIKLIQDKCSCRM--I 213
Query 129 SSKNKFPDSQDRLLLMSGS 147
+SK+ P S +R + + G+
Sbjct 214 ASKDMLPQSTERTVEIHGT 232
> CE09674
Length=397
Score = 37.0 bits (84), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 31/56 (55%), Gaps = 2/56 (3%)
Query 91 PFHMKLLIDPALANSLIGENSQTITKVAEECGCSMHVLSSKNKFPDSQDRLLLMSG 146
P +++L+ + A +LIG N I ++ E+C + + + P S DR+L+ SG
Sbjct 124 PCEVRMLVHQSHAGALIGRNGSKIKELREKCSARLKIFTG--CAPGSTDRVLITSG 177
> Hs6031187
Length=486
Score = 36.2 bits (82), Expect = 0.024, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 37/61 (60%), Gaps = 3/61 (4%)
Query 90 GPFHMKLLIDPALANSLIGENSQTITKVAEECGCSM---HVLSSKNKFPDSQDRLLLMSG 146
G + +K+LI A S+IG+ QTI ++ +E G ++ + SK+ +P + +R+ L+ G
Sbjct 48 GQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKLSKSKDFYPGTTERVCLIQG 107
Query 147 S 147
+
Sbjct 108 T 108
> Hs4505425
Length=510
Score = 35.8 bits (81), Expect = 0.031, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 37/61 (60%), Gaps = 3/61 (4%)
Query 90 GPFHMKLLIDPALANSLIGENSQTITKVAEECGCSM---HVLSSKNKFPDSQDRLLLMSG 146
G + +K+LI A S+IG+ QTI ++ +E G ++ + SK+ +P + +R+ L+ G
Sbjct 48 GQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKLSKSKDFYPGTTERVCLIQG 107
Query 147 S 147
+
Sbjct 108 T 108
> 7296507
Length=386
Score = 35.4 bits (80), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 32/54 (59%), Gaps = 2/54 (3%)
Query 94 MKLLIDPALANSLIGENSQTITKVAEECGCSMHVLSSKNKFPDSQDRLLLMSGS 147
++L++ + SLIG++ I ++ + GCS+ V S P+S +R + +SGS
Sbjct 109 IRLIVPASQCGSLIGKSGSKIKEIRQTTGCSIQVAS--EMLPNSTERAVTLSGS 160
> 7299182
Length=493
Score = 34.3 bits (77), Expect = 0.097, Method: Composition-based stats.
Identities = 14/54 (25%), Positives = 33/54 (61%), Gaps = 0/54 (0%)
Query 94 MKLLIDPALANSLIGENSQTITKVAEECGCSMHVLSSKNKFPDSQDRLLLMSGS 147
MK+L+ + ++IG+ +TI + ++ G + + S + +P + +R+ L++GS
Sbjct 1 MKILVPAVASGAIIGKGGETIASLQKDTGARVKMSKSHDFYPGTTERVCLITGS 54
> At3g04610
Length=577
Score = 32.7 bits (73), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 30/60 (50%), Gaps = 2/60 (3%)
Query 89 PGPFHMKLLIDPALANSLIGENSQTITKVAEECGCSMHVLSSKN--KFPDSQDRLLLMSG 146
P +LL+ + A SLIG+ T+ + E C + VL S++ F DR++ + G
Sbjct 274 PSKVSTRLLVPASQAGSLIGKQGGTVKAIQEASACIVRVLGSEDLPVFALQDDRVVEVVG 333
Score = 31.6 bits (70), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 17/66 (25%), Positives = 31/66 (46%), Gaps = 3/66 (4%)
Query 81 EDERPVSLPGPFHMKLLIDPALANSLIGENSQTITKVAEECGCSMHVLSSKNKFPDSQDR 140
E++R PG ++L+ S+IG I K+ EE + +L P + +R
Sbjct 176 EEKRWPGWPGETVFRMLVPAQKVGSIIGRKGDVIKKIVEETRARIKILDGP---PGTTER 232
Query 141 LLLMSG 146
+++SG
Sbjct 233 AVMVSG 238
> 7302361
Length=499
Score = 32.3 bits (72), Expect = 0.34, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 30/55 (54%), Gaps = 1/55 (1%)
Query 92 FHMKLLIDPALANSLIGENSQTITKVAEECGCSMHVLSSKNKFPDSQDRLLLMSG 146
F ++LLI +LA +IG+ Q I ++ + GC + S N P S DR++ G
Sbjct 91 FDVRLLIHQSLAGCVIGKGGQKIKEIRDRIGCRFLKVFS-NVAPQSTDRVVQTVG 144
> 7302869
Length=797
Score = 31.2 bits (69), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 22/42 (52%), Gaps = 2/42 (4%)
Query 106 LIGENSQTITKVAEECGCSMHVLSSKNKFPDSQDRLLLMSGS 147
+IG+ I K+ ECGC + + KN + DR ++ G+
Sbjct 330 VIGKGGDMIRKIQTECGCKLQFIQGKN--DEMGDRRCVIQGT 369
> Hs22045883
Length=1024
Score = 30.4 bits (67), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 24/97 (24%), Positives = 38/97 (39%), Gaps = 7/97 (7%)
Query 15 ISVLGPLEQVTIAIKAMLPYIHADPNQSQSVHQSYGHGYKLKMPSWAETGVLQEGMNMEA 74
+S GPLE I +H N V + G+ +P W+ +GV
Sbjct 269 LSSCGPLETFKIKYDRKKHQLHVHRN----VASPHPSGHCRPLPGWSTSGVPASAHQPRL 324
Query 75 LKEEPLEDERPVSLPGPFHMKLLIDPALANSLIGENS 111
L++ P LPG + + + P+L + L NS
Sbjct 325 LRQTPFAFREERELPG---LCISLPPSLQSPLFSRNS 358
> At4g26000
Length=495
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 30/65 (46%), Gaps = 3/65 (4%)
Query 82 DERPVSLPGPFHMKLLIDPALANSLIGENSQTITKVAEECGCSMHVLSSKNKFPDSQDRL 141
+ER PG ++++ ++IG I K+ EE + VL P DR+
Sbjct 64 EERWPGWPGDCVFRMIVPVTKVGAIIGRKGDFIKKMCEETRARIKVLDGPVNTP---DRI 120
Query 142 LLMSG 146
+L+SG
Sbjct 121 VLISG 125
> YPR055w
Length=1065
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 19/69 (27%), Positives = 35/69 (50%), Gaps = 7/69 (10%)
Query 75 LKEEPLEDERPVSLPGPFHMKLLIDPALANSLIGENSQTITKVAEECGCSMHVLSSKNKF 134
+K+E E + P+ P F+MK+++DP L L +++ TI + + +SS F
Sbjct 534 VKDESFEQDEPLVPPSVFNMKVILDPFL---LFTQSTSTIVPSV----LTQNTISSLTFF 586
Query 135 PDSQDRLLL 143
D ++ L
Sbjct 587 DDYMNKSFL 595
> At1g48710
Length=1352
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 24/51 (47%), Gaps = 7/51 (13%)
Query 65 VLQEGMNMEALKEEPLEDERPVSLPGPFHMKLL-------IDPALANSLIG 108
+ QEG E LK+ ++D PV P +KL +DP SL+G
Sbjct 1081 ITQEGYAKEVLKKFKMDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVG 1131
> At1g11265
Length=1352
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 24/51 (47%), Gaps = 7/51 (13%)
Query 65 VLQEGMNMEALKEEPLEDERPVSLPGPFHMKLL-------IDPALANSLIG 108
+ QEG E LK+ ++D PV P +KL +DP SL+G
Sbjct 1081 ITQEGYAKEVLKKFKMDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVG 1131
> At1g58140
Length=1320
Score = 28.5 bits (62), Expect = 4.3, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 24/51 (47%), Gaps = 7/51 (13%)
Query 65 VLQEGMNMEALKEEPLEDERPVSLPGPFHMKLL-------IDPALANSLIG 108
+ QEG E LK+ ++D PV P +KL +DP SL+G
Sbjct 1049 ITQEGYAKEVLKKFKMDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVG 1099
> At3g45640
Length=370
Score = 28.5 bits (62), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 19/79 (24%), Positives = 39/79 (49%), Gaps = 6/79 (7%)
Query 6 VPSNPHERIISVLGPLEQVTIAIKAMLPYIHADPNQSQSVHQSYGHGYKLKMPSWAETGV 65
P P ++ S + P+ AI + + DPN+ +V Q+ H Y K+ + +
Sbjct 281 FPRQPLAKLFSHVNPM-----AIDLVDRMLTFDPNRRITVEQALNHQYLAKLHDPNDEPI 335
Query 66 LQEGMNMEALKEEPLEDER 84
Q+ + E +++PL++E+
Sbjct 336 CQKPFSFE-FEQQPLDEEQ 353
> At3g61330
Length=1352
Score = 28.5 bits (62), Expect = 4.7, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 24/51 (47%), Gaps = 7/51 (13%)
Query 65 VLQEGMNMEALKEEPLEDERPVSLPGPFHMKLL-------IDPALANSLIG 108
+ QEG E LK+ ++D PV P +KL +DP SL+G
Sbjct 1081 ITQEGYAKEVLKKFKIDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVG 1131
> At1g21945
Length=1311
Score = 28.5 bits (62), Expect = 4.7, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 24/51 (47%), Gaps = 7/51 (13%)
Query 65 VLQEGMNMEALKEEPLEDERPVSLPGPFHMKLL-------IDPALANSLIG 108
+ QEG E LK+ ++D PV P +KL +DP SL+G
Sbjct 1040 ITQEGYAKEVLKKFKMDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVG 1090
> CE21179
Length=292
Score = 28.5 bits (62), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 69 GMNMEALKEEPLEDERPVSLPGPFHMKLLID 99
G E L ++ +E ++P LP P +M+LL+D
Sbjct 153 GSRYEYLWQDGIEYKKPTRLPAPQYMQLLMD 183
> CE08736
Length=497
Score = 28.5 bits (62), Expect = 5.1, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 11 HERIISVLGPLEQVTIAIKAMLPYIHADPNQSQ 43
HE + V+G +T A K LPY +A N+SQ
Sbjct 332 HEELDKVIGSDRLITTADKNDLPYFNASINESQ 364
> CE05266
Length=589
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Query 94 MKLLIDPALANSLIGENSQTITKVAEECGCSMHVLSSKNKFPDSQDRLLLMSG 146
+ + I P +IG++ TI ++ E+ GC M +L N+ Q + L ++G
Sbjct 135 IDIAIPPNRCGLIIGKSGDTIRQLQEKSGCKM-ILVQDNQSVSDQSKPLRITG 186
> CE22152
Length=244
Score = 28.1 bits (61), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 26/52 (50%), Gaps = 4/52 (7%)
Query 60 WAETGVLQEGMNMEALKEEPLEDERPVSLPGPFHMKLL----IDPALANSLI 107
+ + +LQ + ALK+ P E + LP P MKL+ I LAN L+
Sbjct 40 YGISTLLQYSDRLYALKQRPYEKPLGIFLPSPTAMKLVSRQTISDELANCLL 91
> CE05267
Length=611
Score = 28.1 bits (61), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Query 94 MKLLIDPALANSLIGENSQTITKVAEECGCSMHVLSSKNKFPDSQDRLLLMSG 146
+ + I P +IG++ TI ++ E+ GC M +L N+ Q + L ++G
Sbjct 157 IDIAIPPNRCGLIIGKSGDTIRQLQEKSGCKM-ILVQDNQSVSDQSKPLRITG 208
> CE22218
Length=495
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 17/43 (39%), Positives = 24/43 (55%), Gaps = 2/43 (4%)
Query 1 HPGQLVPSNPHERIISVLGPLEQVTIAIKAMLPYIHADPNQSQ 43
HP V E + V+G VTI+ K+ LPY++A N+SQ
Sbjct 321 HPE--VQDKMQEELDKVVGSDRLVTISDKSSLPYMNAVINESQ 361
> 7302610
Length=1268
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 21/43 (48%), Gaps = 6/43 (13%)
Query 98 IDPALANSLIGENSQTITKVAEECGCSMHVLSSKNKFPDSQDR 140
I P NS+IG + I+ + EECG KFP+S +
Sbjct 642 IPPKYYNSIIGTGGKLISSIMEECG------GVSIKFPNSDSK 678
> Hs18569950
Length=291
Score = 27.7 bits (60), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 33/71 (46%), Gaps = 4/71 (5%)
Query 35 IHADPNQSQSVHQSYGHGYKLKMPSWAETGVLQEGMNMEALKEEPLEDERPVSLPGPFHM 94
+H D N+S V S G +LK G+ Q NM ++E + R PG H
Sbjct 58 VHRD-NESPFVRNSLPVGQELKWDEKTREGIKQHETNMRENQQESILTSR--ECPGAVHR 114
Query 95 KLLIDPALANS 105
K+L + A A+S
Sbjct 115 KVL-EKAAADS 124
Lambda K H
0.313 0.131 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1785281974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40