bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
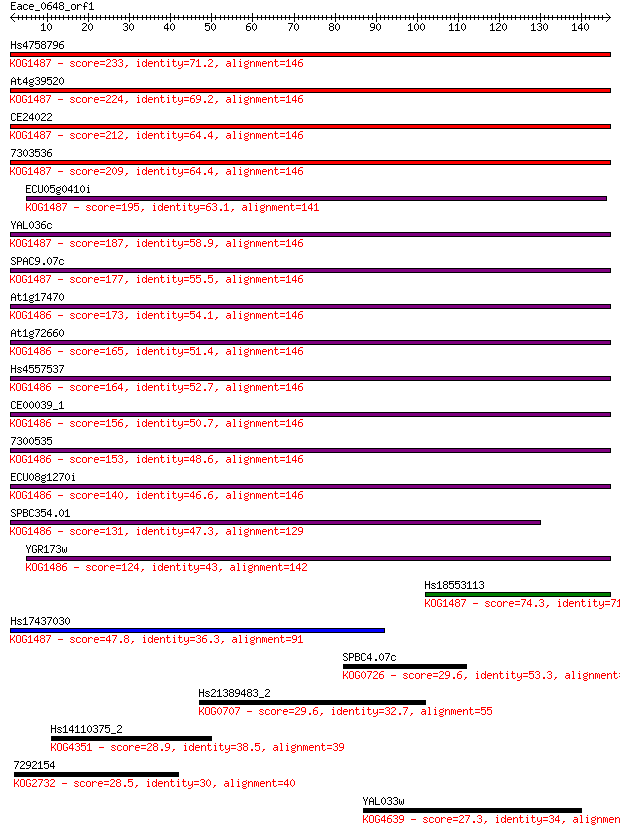
Query= Eace_0648_orf1
Length=146
Score E
Sequences producing significant alignments: (Bits) Value
Hs4758796 233 8e-62
At4g39520 224 4e-59
CE24022 212 2e-55
7303536 209 2e-54
ECU05g0410i 195 2e-50
YAL036c 187 6e-48
SPAC9.07c 177 5e-45
At1g17470 173 9e-44
At1g72660 165 2e-41
Hs4557537 164 5e-41
CE00039_1 156 1e-38
7300535 153 1e-37
ECU08g1270i 140 9e-34
SPBC354.01 131 5e-31
YGR173w 124 4e-29
Hs18553113 74.3 7e-14
Hs17437030 47.8 8e-06
SPBC4.07c 29.6 1.9
Hs21389483_2 29.6 2.2
Hs14110375_2 28.9 4.0
7292154 28.5 4.3
YAL033w 27.3 9.8
> Hs4758796
Length=367
Score = 233 bits (595), Expect = 8e-62, Method: Compositional matrix adjust.
Identities = 104/146 (71%), Positives = 129/146 (88%), Gaps = 0/146 (0%)
Query 1 RCNATADELIDVIEGNRLYVPCIYAINKIDQITIEELDILEQVPHYVPISANLELNLDGL 60
R +ATAD+LIDV+EGNR+Y+PCIY +NKIDQI+IEELDI+ +VPH VPISA+ N D L
Sbjct 222 RSDATADDLIDVVEGNRVYIPCIYVLNKIDQISIEELDIIYKVPHCVPISAHHRWNFDDL 281
Query 61 LEKIWEYLSLVRVYTKPKGQIPDYSAPVILPSTRCKVEDFCMRIHKSLLNEFKQAIVWGR 120
LEKIW+YL LVR+YTKPKGQ+PDY++PV+LP +R VEDFCM+IHK+L+ EFK A+VWG
Sbjct 282 LEKIWDYLKLVRIYTKPKGQLPDYTSPVVLPYSRTTVEDFCMKIHKNLIKEFKYALVWGL 341
Query 121 SVKHNPQKVGKEHQLEDEDVVQIIKR 146
SVKHNPQKVGK+H LEDEDV+QI+K+
Sbjct 342 SVKHNPQKVGKDHTLEDEDVIQIVKK 367
> At4g39520
Length=369
Score = 224 bits (571), Expect = 4e-59, Method: Compositional matrix adjust.
Identities = 101/146 (69%), Positives = 125/146 (85%), Gaps = 0/146 (0%)
Query 1 RCNATADELIDVIEGNRLYVPCIYAINKIDQITIEELDILEQVPHYVPISANLELNLDGL 60
R +ATAD+LIDVIEG+R+Y+PCIYA+NKID IT+EEL+IL+++PHY P+SA+LE NLDGL
Sbjct 223 RYDATADDLIDVIEGSRIYMPCIYAVNKIDSITLEELEILDKLPHYCPVSAHLEWNLDGL 282
Query 61 LEKIWEYLSLVRVYTKPKGQIPDYSAPVILPSTRCKVEDFCMRIHKSLLNEFKQAIVWGR 120
L+KIWEYL L R+YTKPK PDY PVIL S + VEDFC+RIHK +L +FK A+VWG
Sbjct 283 LDKIWEYLDLTRIYTKPKAMNPDYDDPVILSSKKRTVEDFCIRIHKDMLKQFKYALVWGS 342
Query 121 SVKHNPQKVGKEHQLEDEDVVQIIKR 146
S KH PQ+VGKEH+LEDEDVVQI+K+
Sbjct 343 SAKHKPQRVGKEHELEDEDVVQIVKK 368
> CE24022
Length=366
Score = 212 bits (540), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 94/146 (64%), Positives = 124/146 (84%), Gaps = 0/146 (0%)
Query 1 RCNATADELIDVIEGNRLYVPCIYAINKIDQITIEELDILEQVPHYVPISANLELNLDGL 60
R +AT+++LIDVIEGNR+Y+PCIY +NKIDQI+IEELDI+ ++PH VPISA+ + N D L
Sbjct 220 RYDATSEDLIDVIEGNRIYIPCIYVLNKIDQISIEELDIIYRIPHTVPISAHHKWNFDDL 279
Query 61 LEKIWEYLSLVRVYTKPKGQIPDYSAPVILPSTRCKVEDFCMRIHKSLLNEFKQAIVWGR 120
LEK+WEYL+L+R+YTKPKGQ+PDYS P++L + R +ED C +IHKSL +FK A+VWG
Sbjct 280 LEKVWEYLNLIRIYTKPKGQLPDYSQPIVLNAERKSIEDLCTKIHKSLQKDFKCALVWGA 339
Query 121 SVKHNPQKVGKEHQLEDEDVVQIIKR 146
S KHNPQ+VG++H L DEDVVQ+IK+
Sbjct 340 SAKHNPQRVGRDHVLIDEDVVQVIKK 365
> 7303536
Length=368
Score = 209 bits (531), Expect = 2e-54, Method: Compositional matrix adjust.
Identities = 94/146 (64%), Positives = 121/146 (82%), Gaps = 0/146 (0%)
Query 1 RCNATADELIDVIEGNRLYVPCIYAINKIDQITIEELDILEQVPHYVPISANLELNLDGL 60
R +AT+D+LIDVIEGNR+Y+PCIY +NKIDQI+IEELD++ ++PH VPISA+ N D L
Sbjct 222 RYDATSDDLIDVIEGNRIYIPCIYLLNKIDQISIEELDVIYKIPHCVPISAHHHWNFDDL 281
Query 61 LEKIWEYLSLVRVYTKPKGQIPDYSAPVILPSTRCKVEDFCMRIHKSLLNEFKQAIVWGR 120
LE +WEYL L R+YTKPKGQ+PDY++PV+L + R +EDFC ++H+S+ EFK A+VWG
Sbjct 282 LELMWEYLRLQRIYTKPKGQLPDYNSPVVLHNERTSIEDFCNKLHRSIAKEFKYALVWGS 341
Query 121 SVKHNPQKVGKEHQLEDEDVVQIIKR 146
SVKH PQKVG EH L DEDVVQI+K+
Sbjct 342 SVKHQPQKVGIEHVLNDEDVVQIVKK 367
> ECU05g0410i
Length=362
Score = 195 bits (496), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 89/141 (63%), Positives = 107/141 (75%), Gaps = 0/141 (0%)
Query 5 TADELIDVIEGNRLYVPCIYAINKIDQITIEELDILEQVPHYVPISANLELNLDGLLEKI 64
T D+LID IEGNR YVPCIY +NKID I++EELD+L+++PH VP+ AN N D LL K+
Sbjct 220 TIDDLIDAIEGNRKYVPCIYVLNKIDSISVEELDLLDKIPHSVPVCANFGWNFDSLLSKM 279
Query 65 WEYLSLVRVYTKPKGQIPDYSAPVILPSTRCKVEDFCMRIHKSLLNEFKQAIVWGRSVKH 124
W YL LVR+Y KPKG+ DY PVIL S + V DFC IH+ +L +FK A+VWG SVKH
Sbjct 280 WSYLDLVRIYPKPKGEPIDYEEPVILRSNKRSVADFCNAIHRGILAKFKHALVWGSSVKH 339
Query 125 NPQKVGKEHQLEDEDVVQIIK 145
NPQKVGKEH L D DVVQI+K
Sbjct 340 NPQKVGKEHILNDSDVVQIVK 360
> YAL036c
Length=369
Score = 187 bits (475), Expect = 6e-48, Method: Compositional matrix adjust.
Identities = 86/147 (58%), Positives = 119/147 (80%), Gaps = 1/147 (0%)
Query 1 RCNATADELIDVIEGN-RLYVPCIYAINKIDQITIEELDILEQVPHYVPISANLELNLDG 59
RC+AT D+LIDV+E + R Y+P IY +NKID ++IEEL++L ++P+ VPIS+ + NLD
Sbjct 223 RCDATVDDLIDVLEASSRRYMPAIYVLNKIDSLSIEELELLYRIPNAVPISSGQDWNLDE 282
Query 60 LLEKIWEYLSLVRVYTKPKGQIPDYSAPVILPSTRCKVEDFCMRIHKSLLNEFKQAIVWG 119
LL+ +W+ L+LVR+YTKPKGQIPD++ PV+L S RC V+DFC +IHKSL+++F+ A+V+G
Sbjct 283 LLQVMWDRLNLVRIYTKPKGQIPDFTDPVVLRSDRCSVKDFCNQIHKSLVDDFRNALVYG 342
Query 120 RSVKHNPQKVGKEHQLEDEDVVQIIKR 146
SVKH PQ VG H LEDEDVV I+K+
Sbjct 343 SSVKHQPQYVGLSHILEDEDVVTILKK 369
> SPAC9.07c
Length=366
Score = 177 bits (450), Expect = 5e-45, Method: Compositional matrix adjust.
Identities = 81/146 (55%), Positives = 110/146 (75%), Gaps = 0/146 (0%)
Query 1 RCNATADELIDVIEGNRLYVPCIYAINKIDQITIEELDILEQVPHYVPISANLELNLDGL 60
RC+AT D+LIDV+EGNR+Y+P +Y +NKID I+IEELD+++++P+ VPI N N+D L
Sbjct 221 RCDATIDDLIDVLEGNRVYIPALYVLNKIDSISIEELDLIDRIPNAVPICGNRGWNIDEL 280
Query 61 LEKIWEYLSLVRVYTKPKGQIPDYSAPVILPSTRCKVEDFCMRIHKSLLNEFKQAIVWGR 120
E +W+YL+LVRVYT+P+G PDYS PVIL + VEDFC IH S+ ++FK A VWG+
Sbjct 281 KETMWDYLNLVRVYTRPRGLEPDYSEPVILRTGHSTVEDFCNNIHSSIKSQFKHAYVWGK 340
Query 121 SVKHNPQKVGKEHQLEDEDVVQIIKR 146
SV + +VG H L DEDVV I+K+
Sbjct 341 SVPYPGMRVGLSHVLLDEDVVTIVKK 366
> At1g17470
Length=399
Score = 173 bits (439), Expect = 9e-44, Method: Compositional matrix adjust.
Identities = 79/148 (53%), Positives = 107/148 (72%), Gaps = 2/148 (1%)
Query 1 RCNATADELIDVIEGNRLYVPCIYAINKIDQITIEELDILEQVPHYVPISANLELNLDGL 60
R NAT D+ IDVIEGNR Y+ C+Y NKID + I+++D L + P+ + IS NL+LNLD L
Sbjct 220 RENATVDDFIDVIEGNRKYIKCVYVYNKIDVVGIDDVDRLSRQPNSIVISCNLKLNLDRL 279
Query 61 LEKIWEYLSLVRVYTKPKGQIPDYSAPVILPSTR--CKVEDFCMRIHKSLLNEFKQAIVW 118
L ++W+ + LVRVY+KP+GQ PD+ P +L S R C VEDFC +H++L+ + K A+VW
Sbjct 280 LARMWDEMGLVRVYSKPQGQQPDFDEPFVLSSDRGGCTVEDFCNHVHRTLVKDMKYALVW 339
Query 119 GRSVKHNPQKVGKEHQLEDEDVVQIIKR 146
G S +HNPQ G LEDEDVVQI+K+
Sbjct 340 GTSTRHNPQNCGLSQHLEDEDVVQIVKK 367
> At1g72660
Length=399
Score = 165 bits (418), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 75/148 (50%), Positives = 107/148 (72%), Gaps = 2/148 (1%)
Query 1 RCNATADELIDVIEGNRLYVPCIYAINKIDQITIEELDILEQVPHYVPISANLELNLDGL 60
R +AT D+ IDV+EGNR Y+ C+Y NKID + I+++D L + P+ + IS NL+LNLD L
Sbjct 220 REDATVDDFIDVVEGNRKYIKCVYVYNKIDVVGIDDVDRLARQPNSIVISCNLKLNLDRL 279
Query 61 LEKIWEYLSLVRVYTKPKGQIPDYSAPVILPSTR--CKVEDFCMRIHKSLLNEFKQAIVW 118
L ++W+ + LVRVY+KP+ Q PD+ P +L + R C VEDFC ++H++L+ + K A+VW
Sbjct 280 LARMWDEMGLVRVYSKPQSQQPDFDEPFVLSADRGGCTVEDFCNQVHRTLVKDMKYALVW 339
Query 119 GRSVKHNPQKVGKEHQLEDEDVVQIIKR 146
G S +H PQ G H LEDEDVVQI+K+
Sbjct 340 GTSARHYPQHCGLFHHLEDEDVVQIVKK 367
> Hs4557537
Length=364
Score = 164 bits (415), Expect = 5e-41, Method: Compositional matrix adjust.
Identities = 77/146 (52%), Positives = 107/146 (73%), Gaps = 1/146 (0%)
Query 1 RCNATADELIDVIEGNRLYVPCIYAINKIDQITIEELDILEQVPHYVPISANLELNLDGL 60
R + + DE IDVI GNR+Y+PC+Y NKIDQI++EE+D L + P+ V IS ++LNLD L
Sbjct 220 REDCSPDEFIDVIVGNRVYMPCLYVYNKIDQISMEEVDRLARKPNSVVISCGMKLNLDYL 279
Query 61 LEKIWEYLSLVRVYTKPKGQIPDYSAPVILPSTRCKVEDFCMRIHKSLLNEFKQAIVWGR 120
LE +WEYL+L +YTK +GQ PD++ +IL VE C RIH+SL ++FK A+VWG
Sbjct 280 LEMLWEYLALTCIYTKKRGQRPDFTDAIILRKG-ASVEHVCHRIHRSLASQFKYALVWGT 338
Query 121 SVKHNPQKVGKEHQLEDEDVVQIIKR 146
S K++PQ+VG H +E EDV+QI+K+
Sbjct 339 STKYSPQRVGLTHTMEHEDVIQIVKK 364
> CE00039_1
Length=408
Score = 156 bits (394), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 74/147 (50%), Positives = 103/147 (70%), Gaps = 1/147 (0%)
Query 1 RCNATADELIDVIEGNRLYVPCIYAINKIDQITIEELDILEQVPHYVPISANLELNLDGL 60
R + T DE IDVI+GNR+Y+ C+Y NK+DQI+IEE+D L ++PH+V IS + LN+D L
Sbjct 220 REDCTVDEFIDVIQGNRVYMTCLYVYNKVDQISIEEIDRLARMPHHVVISCEMNLNMDYL 279
Query 61 LEKIWEYLSLVRVYTKPKGQIPDYSAP-VILPSTRCKVEDFCMRIHKSLLNEFKQAIVWG 119
LEK+WEYL+LVRVYTK G PD I+ +E C +H+S+ + + AIVWG
Sbjct 280 LEKMWEYLALVRVYTKKPGNAPDLGPEDGIILRGGATIEHCCHALHRSIAAQLRYAIVWG 339
Query 120 RSVKHNPQKVGKEHQLEDEDVVQIIKR 146
S K +PQ+VG H+L+ EDV+QI++R
Sbjct 340 TSTKFSPQRVGLHHKLDHEDVIQIMRR 366
> 7300535
Length=363
Score = 153 bits (386), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 71/146 (48%), Positives = 102/146 (69%), Gaps = 1/146 (0%)
Query 1 RCNATADELIDVIEGNRLYVPCIYAINKIDQITIEELDILEQVPHYVPISANLELNLDGL 60
R + T DE IDV+ NR+Y+PC+Y NKIDQI+IEE+D L + P+ + +S N++LNLD +
Sbjct 219 REDCTEDEFIDVVTANRVYLPCLYVYNKIDQISIEEVDRLARQPNSIVVSCNMKLNLDYM 278
Query 61 LEKIWEYLSLVRVYTKPKGQIPDYSAPVILPSTRCKVEDFCMRIHKSLLNEFKQAIVWGR 120
+E +WE L L+RVYTK G PD+ +IL VE C IH++L +FK A+VWG
Sbjct 279 MEALWEALQLIRVYTKKPGAPPDFDDGLILRKG-VSVEHVCHAIHRTLAAQFKYALVWGT 337
Query 121 SVKHNPQKVGKEHQLEDEDVVQIIKR 146
S K++PQ+VG H + DEDV+Q++K+
Sbjct 338 STKYSPQRVGIAHVMADEDVIQVVKK 363
> ECU08g1270i
Length=362
Score = 140 bits (353), Expect = 9e-34, Method: Compositional matrix adjust.
Identities = 68/146 (46%), Positives = 93/146 (63%), Gaps = 1/146 (0%)
Query 1 RCNATADELIDVIEGNRLYVPCIYAINKIDQITIEELDILEQVPHYVPISANLELNLDGL 60
R + + D++ID+I G+ +YV C+Y NKID++++E+ + + + P+ IS N+ L
Sbjct 218 REDVSDDDIIDLISGSAVYVDCLYVYNKIDELSLEDFNRVAEQPNSTVISCRRNWNISRL 277
Query 61 LEKIWEYLSLVRVYTKPKGQIPDYSAPVILPSTRCKVEDFCMRIHKSLLNEFKQAIVWGR 120
LE IW+ L L RVYTK KG P PV++ V+D C RIHK L FK A+VWG
Sbjct 278 LEDIWDKLKLTRVYTKKKGAFPSLDDPVVIRKG-GTVKDLCSRIHKDFLLSFKHALVWGT 336
Query 121 SVKHNPQKVGKEHQLEDEDVVQIIKR 146
S KH+PQ+VG H LEDEDVVQI R
Sbjct 337 SAKHSPQRVGLGHTLEDEDVVQIFLR 362
> SPBC354.01
Length=346
Score = 131 bits (329), Expect = 5e-31, Method: Compositional matrix adjust.
Identities = 61/129 (47%), Positives = 90/129 (69%), Gaps = 1/129 (0%)
Query 1 RCNATADELIDVIEGNRLYVPCIYAINKIDQITIEELDILEQVPHYVPISANLELNLDGL 60
R + T D+ ID++ GNR Y+ C+Y +KID +++EE+D L ++P V IS N++LNLD L
Sbjct 219 REDITVDDFIDLVMGNRRYINCLYCYSKIDAVSLEEVDRLARLPKSVVISCNMKLNLDFL 278
Query 61 LEKIWEYLSLVRVYTKPKGQIPDYSAPVILPSTRCKVEDFCMRIHKSLLNEFKQAIVWGR 120
E+IWE L+L R+YTK KG++PD+S +I+ +E C RIH++L + K A+VWG
Sbjct 279 KERIWEELNLYRIYTKRKGEMPDFSEALIVRKG-STIEQVCNRIHRTLAEQLKYALVWGT 337
Query 121 SVKHNPQKV 129
S KH+PQ V
Sbjct 338 SAKHSPQVV 346
> YGR173w
Length=368
Score = 124 bits (312), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 61/143 (42%), Positives = 94/143 (65%), Gaps = 2/143 (1%)
Query 5 TADELIDVI-EGNRLYVPCIYAINKIDQITIEELDILEQVPHYVPISANLELNLDGLLEK 63
T D+ IDVI E +R YV C+Y NKID +++EE+D L + P+ V +S ++L L ++E+
Sbjct 227 TIDDFIDVINEQHRNYVKCLYVYNKIDAVSLEEVDKLAREPNTVVMSCEMDLGLQDVVEE 286
Query 64 IWEYLSLVRVYTKPKGQIPDYSAPVILPSTRCKVEDFCMRIHKSLLNEFKQAIVWGRSVK 123
IW L+L RVYTK +G P + P+++ + + D C IH+ ++FK A+VWG S K
Sbjct 287 IWYQLNLSRVYTKKRGVRPVFDDPLVVRNNST-IGDLCHGIHRDFKDKFKYALVWGSSAK 345
Query 124 HNPQKVGKEHQLEDEDVVQIIKR 146
H+PQK G H+++DEDVV + +
Sbjct 346 HSPQKCGLNHRIDDEDVVSLFAK 368
> Hs18553113
Length=94
Score = 74.3 bits (181), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 32/45 (71%), Positives = 41/45 (91%), Gaps = 0/45 (0%)
Query 102 MRIHKSLLNEFKQAIVWGRSVKHNPQKVGKEHQLEDEDVVQIIKR 146
M+IHK+L+ EFK A+VWG SVKHNPQKVGK+H LED+DV+QI+K+
Sbjct 1 MKIHKNLIKEFKYALVWGLSVKHNPQKVGKDHTLEDKDVIQIVKK 45
> Hs17437030
Length=104
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 33/91 (36%), Positives = 51/91 (56%), Gaps = 9/91 (9%)
Query 1 RCNATADELIDVIEGNRLYVPCIYAINKIDQITIEELDILEQVPHYVPISANLELNLDGL 60
+ +ATA+ LI V++ N + V + N+I+ I+ +LDI+ V H IS +
Sbjct 22 QSDATANGLIGVVKENAV-VFSVSGRNQIEHISTNKLDIVHTVTHQRLISTPKQC----- 75
Query 61 LEKIWEYLSLVRVYTKPKGQIPDYSAPVILP 91
I E LSLV+VYT PKGQ + ++ V+L
Sbjct 76 ---IGEDLSLVKVYTNPKGQPQNCTSLVVLS 103
> SPBC4.07c
Length=448
Score = 29.6 bits (65), Expect = 1.9, Method: Composition-based stats.
Identities = 16/33 (48%), Positives = 21/33 (63%), Gaps = 3/33 (9%)
Query 82 PDYSA--PVILPSTRCKVEDFCM-RIHKSLLNE 111
PD SA P ++P+TRC++ M RIH LL E
Sbjct 50 PDASAKLPTVIPTTRCRLRLLKMQRIHDHLLME 82
> Hs21389483_2
Length=385
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 25/56 (44%), Gaps = 1/56 (1%)
Query 47 VPISANLELNLDGLLEKIWEYLSLVRVYTKPK-GQIPDYSAPVILPSTRCKVEDFC 101
VP L+ LE++W+ L Y KP G P+ S L S +C + FC
Sbjct 254 VPAPLTSGLHYYTTLEELWKSFDLCEDYFKPPFGPYPEKSGKDSLVSMKCSLFRFC 309
> Hs14110375_2
Length=666
Score = 28.9 bits (63), Expect = 4.0, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 23/44 (52%), Gaps = 5/44 (11%)
Query 11 DVIEGNRLYVPCIYAINKIDQITIEELDI-----LEQVPHYVPI 49
+V LY+P + + D ++ E LDI LE+VPH P+
Sbjct 240 NVASERELYIPSVDLLTAQDLLSFELLDINIVQELERVPHNTPV 283
> 7292154
Length=431
Score = 28.5 bits (62), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 12/40 (30%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 2 CNATADELIDVIEGNRLYVPCIYAINKIDQITIEELDILE 41
C + A EL + EG R + C+ + +K + + +LD L+
Sbjct 381 CESFATELHEGSEGKRTRLVCVPSFSKTQSVAVVDLDTLD 420
> YAL033w
Length=173
Score = 27.3 bits (59), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 32/57 (56%), Gaps = 4/57 (7%)
Query 87 PVILPSTRCKVEDFCMRIHKSLLNEFKQAIVWGRS----VKHNPQKVGKEHQLEDED 139
PV + T K+E F MR + +LN K + S + ++ +K+G+E++ E+ED
Sbjct 116 PVKVSGTIKKIEQFAMRRNSKILNIIKCSQSSHLSDNDFIINDFKKIGRENENENED 172
Lambda K H
0.320 0.139 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1748847648
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40