bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
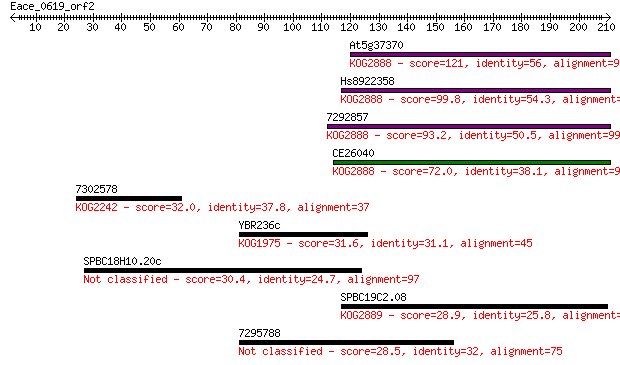
Query= Eace_0619_orf2
Length=210
Score E
Sequences producing significant alignments: (Bits) Value
At5g37370 121 1e-27
Hs8922358 99.8 4e-21
7292857 93.2 3e-19
CE26040 72.0 9e-13
7302578 32.0 0.89
YBR236c 31.6 1.3
SPBC18H10.20c 30.4 2.7
SPBC19C2.08 28.9 8.5
7295788 28.5 9.1
> At5g37370
Length=393
Score = 121 bits (303), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 51/91 (56%), Positives = 67/91 (73%), Gaps = 0/91 (0%)
Query 120 VNQLLRANILSSEYFKSLHQLKSFHEVVAEVAAYADHAEPYCSGSTRAPSTLFCCLYKLF 179
+ ++L NILSS+YFK L+ LK++HEV+ E+ +H EP+ G+ R PST +C LYK F
Sbjct 15 LEKVLSMNILSSDYFKELYGLKTYHEVIDEIYNQVNHVEPWMGGNCRGPSTAYCLLYKFF 74
Query 180 TLKLTDKQMHMLLNHRESPYVRCTGFLYLRY 210
T+KLT KQMH LL H +SPY+R GFLYLRY
Sbjct 75 TMKLTVKQMHGLLKHTDSPYIRAVGFLYLRY 105
> Hs8922358
Length=546
Score = 99.8 bits (247), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 51/113 (45%), Positives = 67/113 (59%), Gaps = 19/113 (16%)
Query 117 TYNVNQLLRANILSSEYFK-SLHQLKSFHEVVAEVAAYADHAEPYCSGSTRA-------- 167
T N+N ++ NILSS YFK L++LK++HEVV E+ H EP+ GS +
Sbjct 55 TMNLNPMILTNILSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTGMCG 114
Query 168 ----------PSTLFCCLYKLFTLKLTDKQMHMLLNHRESPYVRCTGFLYLRY 210
ST FC LYKLFTLKLT KQ+ L+ H +SPY+R GF+Y+RY
Sbjct 115 GVRGVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHTDSPYIRALGFMYIRY 167
> 7292857
Length=398
Score = 93.2 bits (230), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 50/118 (42%), Positives = 69/118 (58%), Gaps = 19/118 (16%)
Query 112 MTDSTTYNVNQLLRANILSSEYFK-SLHQLKSFHEVVAEVAAYADHAEPYCSGSTRAP-- 168
+ ++ N+N L+ ANI SS YFK L +LK++HEVV E+ H EP+ GS +
Sbjct 27 WGNESSMNLNALILANIQSSSYFKVHLFKLKTYHEVVDEIYYQVKHMEPWERGSRKTSGQ 86
Query 169 ----------------STLFCCLYKLFTLKLTDKQMHMLLNHRESPYVRCTGFLYLRY 210
ST +C LYKL+TL+LT KQ++ LLNH +S Y+R GF+YLRY
Sbjct 87 TGMCGGVRGVGAGGIVSTAYCLLYKLYTLRLTRKQINGLLNHTDSAYIRALGFMYLRY 144
> CE26040
Length=327
Score = 72.0 bits (175), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 37/116 (31%), Positives = 62/116 (53%), Gaps = 19/116 (16%)
Query 114 DSTTYNVNQLLRANILSSEYFKS-LHQLKSFHEVVAEVAAYADHAEPYCSGSTR------ 166
+ T N+N L+ NI S Y+K+ L ++ +F ++ ++ H EP+ G+ R
Sbjct 60 NKVTMNLNTLVLENIRESYYYKNNLVEIDNFQTLIEQIFYQVKHLEPWEKGTRRLQGMTG 119
Query 167 ------------APSTLFCCLYKLFTLKLTDKQMHMLLNHRESPYVRCTGFLYLRY 210
S+ +C LY+LF LK++ KQ+ +LN R+S Y+R GF+Y+RY
Sbjct 120 MCGGVRGVGAGGVVSSAYCLLYRLFNLKISRKQLISMLNSRQSVYIRGLGFMYIRY 175
> 7302578
Length=795
Score = 32.0 bits (71), Expect = 0.89, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 24 QRETTCVASAAAPVAVAPLAAAPATDVAAAENDKEDE 60
QR T ++ + +PV AP+AA P D E ED+
Sbjct 58 QRRTRSMSRSPSPVQAAPVAAEPVLDTLEEEEQPEDK 94
> YBR236c
Length=436
Score = 31.6 bits (70), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 81 RKYKSFTQQMGVLQQRAIAADEETQKKNMVQMTDSTTYNVNQLLR 125
R+++ + Q+ + +QRA EE K++ ++MT + + NV+Q++R
Sbjct 79 RRHERYDQEERLRKQRAQKLREEQLKRHEIEMTANRSINVDQIVR 123
> SPBC18H10.20c
Length=361
Score = 30.4 bits (67), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 24/101 (23%), Positives = 41/101 (40%), Gaps = 4/101 (3%)
Query 27 TTCVASAAAPVAVAPLAAA--PATDVAAAENDKEDEEEAIARCHLHTKPQLSCKFCRKYK 84
T VA+ P + P AA T A+ND D + L Q SC+ C +++
Sbjct 180 TNLVANITLPSTLHPHGAALMEVTMTGFAQNDGNDWKINRVTWRLEEHMQFSCQPCERHR 239
Query 85 SFTQQMGVLQQRAIAADEETQKKNMV--QMTDSTTYNVNQL 123
+ + ++R ++ + + QM ST N + L
Sbjct 240 DLVKPRPIEEKRILSTQDLQSGWKFIDNQMFLSTQINTSSL 280
> SPBC19C2.08
Length=210
Score = 28.9 bits (63), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 40/95 (42%), Gaps = 7/95 (7%)
Query 117 TYNVNQLLRANILSSEYFKSLHQLKSFHEVVAEVAAYADHAEPYCSG--STRAPSTLFCC 174
T+ + ++LR I+ S Y+K + F + A E Y G + P+ C
Sbjct 22 TFLIGKILRERIVDSIYWKE----QCFGLNACSLVDRAVRLE-YIGGQYGNQRPTEFICL 76
Query 175 LYKLFTLKLTDKQMHMLLNHRESPYVRCTGFLYLR 209
LYKL + + + L+ E Y+R Y+R
Sbjct 77 LYKLLQIAPEKEIIQQYLSIPEFKYLRALAAFYVR 111
> 7295788
Length=807
Score = 28.5 bits (62), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 43/85 (50%), Gaps = 19/85 (22%)
Query 81 RKYKSFTQQMGVLQQRAIAADEETQ---------KKNMVQMTDSTTYNVNQLLRANILSS 131
RK+K+ ++ VLQQ A EET+ +KN+ + + S R+N L
Sbjct 85 RKFKTLKEEQDVLQQDYDAKVEETRSLKDKLTAAEKNLAEKSAS---------RSNQLHD 135
Query 132 EYFKSLHQLKSFH-EVVAEVAAYAD 155
++F L+ L+ + E++ ++A Y D
Sbjct 136 DFFARLNALELENCELLQKLAEYED 160
Lambda K H
0.322 0.131 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3790221436
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40