bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0612_orf1
Length=167
Score E
Sequences producing significant alignments: (Bits) Value
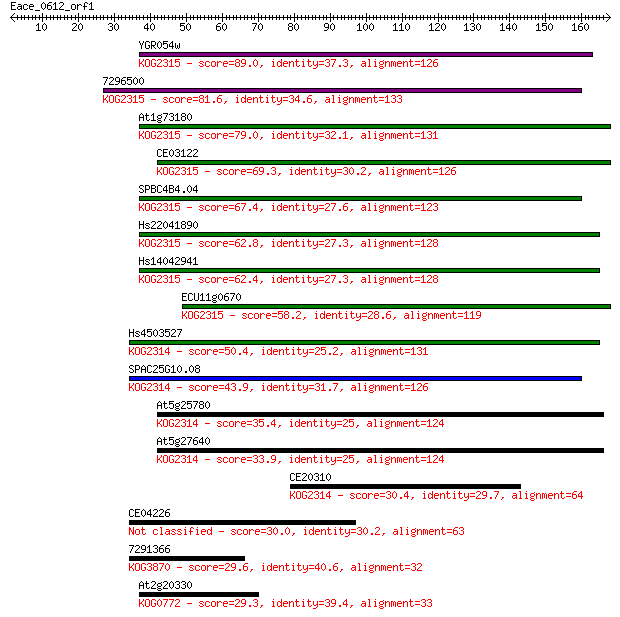
YGR054w 89.0 4e-18
7296500 81.6 6e-16
At1g73180 79.0 5e-15
CE03122 69.3 3e-12
SPBC4B4.04 67.4 1e-11
Hs22041890 62.8 3e-10
Hs14042941 62.4 4e-10
ECU11g0670 58.2 8e-09
Hs4503527 50.4 2e-06
SPAC25G10.08 43.9 1e-04
At5g25780 35.4 0.055
At5g27640 33.9 0.16
CE20310 30.4 1.9
CE04226 30.0 2.4
7291366 29.6 3.2
At2g20330 29.3 3.5
> YGR054w
Length=642
Score = 89.0 bits (219), Expect = 4e-18, Method: Composition-based stats.
Identities = 47/127 (37%), Positives = 68/127 (53%), Gaps = 5/127 (3%)
Query 37 VQDVTWSPTRDEFVLIEGKSPSDIVLLDRQGRTQFVFPRRYRNFVSFCPFGQMAVVAGFG 96
V D TWSPT +F +I G P+ I D +G P++ +N + F P G ++AGFG
Sbjct 294 VHDFTWSPTSRQFGVIAGYMPATISFFDLRGNVVHSLPQQAKNTMLFSPSGHYILIAGFG 353
Query 97 NLAGEVSFYYR-DKASQGMQHISEFKAPCTVLALWAPCGSYFATASTYPRMKVDNCLSVL 155
NL G V R DK + +S+F A T + W+P G + TA+T PR++VDN + +
Sbjct 354 NLQGSVEILDRLDK----FKCVSKFDATNTSVCKWSPGGEFIMTATTSPRLRVDNGVKIW 409
Query 156 DYRGESV 162
G V
Sbjct 410 HVSGSLV 416
> 7296500
Length=638
Score = 81.6 bits (200), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 46/133 (34%), Positives = 63/133 (47%), Gaps = 4/133 (3%)
Query 27 CCVRRISKKIVQDVTWSPTRDEFVLIEGKSPSDIVLLDRQGRTQFVFPRRYRNFVSFCPF 86
C V + V V WSP EFV++ G PS L + + F F RN F PF
Sbjct 282 CSVPLSKEGPVHCVKWSPKATEFVVVYGFMPSKAALYNLKCDVVFDFGEGPRNCAYFNPF 341
Query 87 GQMAVVAGFGNLAGEVSFYYRDKASQGMQHISEFKAPCTVLALWAPCGSYFATASTYPRM 146
G + +AGFGNL G V + K + ++ K T + W P G +F TA+T PR+
Sbjct 342 GNLIALAGFGNLPGAVEVWDVSK----REKLANLKCADTTVFEWHPNGEWFVTATTAPRL 397
Query 147 KVDNCLSVLDYRG 159
++ N V Y G
Sbjct 398 RISNGFKVYHYSG 410
> At1g73180
Length=513
Score = 79.0 bits (193), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 42/131 (32%), Positives = 72/131 (54%), Gaps = 4/131 (3%)
Query 37 VQDVTWSPTRDEFVLIEGKSPSDIVLLDRQGRTQFVFPRRYRNFVSFCPFGQMAVVAGFG 96
V DV WS + EF ++ G P+ + + D+ + N + + P G++ VAGFG
Sbjct 275 VHDVQWSFSGSEFAVVYGFMPACVTIFDKNCKPLMELGEGPYNTLRWNPKGRVLCVAGFG 334
Query 97 NLAGEVSFYYRDKASQGMQHISEFKAPCTVLALWAPCGSYFATASTYPRMKVDNCLSVLD 156
NL G+++F+ D ++ + + KA +V + W+P G YF TAST PR ++DN + + +
Sbjct 335 NLPGDMAFW--DVVNK--KQLGSNKAEWSVTSEWSPDGRYFLTASTAPRRQIDNGMKIFN 390
Query 157 YRGESVQRLQF 167
Y G+ + F
Sbjct 391 YDGKRYYKKMF 401
> CE03122
Length=618
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 38/127 (29%), Positives = 61/127 (48%), Gaps = 5/127 (3%)
Query 42 WSPTRDEFVLIEGKSPSDIVLLDRQGRTQFVFPRRYRNFVSFCPFGQMAVVAGFGNLA-G 100
W+P EF + G P+ + + +G F RN V + FG + ++ GFGN+A G
Sbjct 332 WNPNGREFAVCYGYMPAKVTFYNPRGVPIFDTIEGPRNDVFYNAFGNIVLICGFGNIAKG 391
Query 101 EVSFYYRDKASQGMQHISEFKAPCTVLALWAPCGSYFATASTYPRMKVDNCLSVLDYRGE 160
++ F+ + + I + P T L WAP G +F T +T PR+++DN Y G
Sbjct 392 KMEFW----DVETKKEIISIEVPNTTLFDWAPDGQHFVTCTTAPRLRIDNSYRFWHYTGR 447
Query 161 SVQRLQF 167
+ F
Sbjct 448 MLAETHF 454
> SPBC4B4.04
Length=576
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 34/123 (27%), Positives = 59/123 (47%), Gaps = 3/123 (2%)
Query 37 VQDVTWSPTRDEFVLIEGKSPSDIVLLDRQGRTQFVFPRRYRNFVSFCPFGQMAVVAGFG 96
+ DV W+ EF ++ G P+ + D + + P RN + F P + ++AGFG
Sbjct 271 IHDVCWNADSKEFGIVYGYMPAKTAIFDNRANVVSIIPPAPRNTLIFSPNSRYILLAGFG 330
Query 97 NLAGEVSFYYRDKASQGMQHISEFKAPCTVLALWAPCGSYFATASTYPRMKVDNCLSVLD 156
NL G + + A+ M+ I+ +A ++P + TA T PR++VDN + +
Sbjct 331 NLQGSIDIF---DAANNMKKITTVEAANCTYCEFSPDSQFLLTAVTSPRLRVDNSIKIWH 387
Query 157 YRG 159
G
Sbjct 388 ITG 390
> Hs22041890
Length=458
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 35/128 (27%), Positives = 61/128 (47%), Gaps = 4/128 (3%)
Query 37 VQDVTWSPTRDEFVLIEGKSPSDIVLLDRQGRTQFVFPRRYRNFVSFCPFGQMAVVAGFG 96
+ DV W+ + EF + G P+ + + + F F RN + P G + V+AGFG
Sbjct 149 IYDVVWNSSSTEFCAVYGFMPAKATIFNLKCDPVFDFGTGPRNAAYYSPHGHILVLAGFG 208
Query 97 NLAGEVSFYYRDKASQGMQHISEFKAPCTVLALWAPCGSYFATASTYPRMKVDNCLSVLD 156
NL G++ + + + IS+ A + W P G + TA+ PR++V+N +
Sbjct 209 NLRGQMEVW----DVKNYKLISKPVASDSTYFAWCPDGEHILTATCAPRLRVNNGYKIWH 264
Query 157 YRGESVQR 164
Y G + +
Sbjct 265 YTGSILHK 272
> Hs14042941
Length=609
Score = 62.4 bits (150), Expect = 4e-10, Method: Composition-based stats.
Identities = 35/128 (27%), Positives = 61/128 (47%), Gaps = 4/128 (3%)
Query 37 VQDVTWSPTRDEFVLIEGKSPSDIVLLDRQGRTQFVFPRRYRNFVSFCPFGQMAVVAGFG 96
+ DV W+ + EF + G P+ + + + F F RN + P G + V+AGFG
Sbjct 300 IYDVVWNSSSTEFCAVYGFMPAKATIFNLKCDPVFDFGTGPRNAAYYSPHGHILVLAGFG 359
Query 97 NLAGEVSFYYRDKASQGMQHISEFKAPCTVLALWAPCGSYFATASTYPRMKVDNCLSVLD 156
NL G++ + + + IS+ A + W P G + TA+ PR++V+N +
Sbjct 360 NLRGQMEVW----DVKNYKLISKPVASDSTYFAWCPDGEHILTATCAPRLRVNNGYKIWH 415
Query 157 YRGESVQR 164
Y G + +
Sbjct 416 YTGSILHK 423
> ECU11g0670
Length=496
Score = 58.2 bits (139), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 34/119 (28%), Positives = 56/119 (47%), Gaps = 5/119 (4%)
Query 49 FVLIEGKSPSDIVLLDRQGRTQFVFPRRYRNFVSFCPFGQMAVVAGFGNLAGEVSFYYRD 108
F + G PS + + G+ ++ FP RN + F +AV AGF NL+G++ ++
Sbjct 272 FAVCHGPQPSSVSVHGLDGKLRYNFPEGTRNRIFFNQHENIAVFAGFDNLSGDIEVFH-- 329
Query 109 KASQGMQHISEFKAPCTVLALWAPCGSYFATASTYPRMKVDNCLSVLDYRGESVQRLQF 167
+ ++ F L W GSYF ++T + DN +++ DY G V QF
Sbjct 330 --VPSRKLMARFNVLGASLVDWKESGSYFYVSTT-SYFQEDNKITLYDYYGRKVDERQF 385
> Hs4503527
Length=873
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/136 (24%), Positives = 65/136 (47%), Gaps = 9/136 (6%)
Query 34 KKIVQDVTWSPTRDEFVLIEGKSPSDIVL---LDRQGRTQFV--FPRRYRNFVSFCPFGQ 88
K+ + W P +F ++ G++P V + G+ + + F ++ N + + P GQ
Sbjct 562 KETIIAFAWEPNGSKFAVLHGEAPRISVSFYHVKNNGKIELIKMFDKQQANTIFWSPQGQ 621
Query 89 MAVVAGFGNLAGEVSFYYRDKASQGMQHISEFKAPCTVLALWAPCGSYFATASTYPRMKV 148
V+AG ++ G ++F D + + +I+E V W P G Y T+ ++ KV
Sbjct 622 FVVLAGLRSMNGALAFV--DTSDCTVMNIAEHYMASDVE--WDPTGRYVVTSVSWWSHKV 677
Query 149 DNCLSVLDYRGESVQR 164
DN + ++G +Q+
Sbjct 678 DNAYWLWTFQGRLLQK 693
> SPAC25G10.08
Length=725
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 40/154 (25%), Positives = 60/154 (38%), Gaps = 31/154 (20%)
Query 34 KKIVQDVTWSPTRDEFVLIEGKSPSDIVLLDRQGRTQFVF---------PRRYRNFVSF- 83
K +V + W P D F +I S +D VL +T F P +R+ ++F
Sbjct 456 KDVVLNFAWEPKSDRFAII---SANDQVLNSTNVKTNLSFYGFEQKKNTPSTFRHIITFD 512
Query 84 ---------CPFGQMAVVAGFGNLAG-EVSFYYRD--------KASQGMQHISEFKAPCT 125
P G+ V A G+ ++ FY D A +Q I +
Sbjct 513 KKTCNSLFMAPKGRFMVAATLGSSTQYDLEFYDLDFDTEKKEPDALANVQQIGSAEHFGM 572
Query 126 VLALWAPCGSYFATASTYPRMKVDNCLSVLDYRG 159
W P G Y T+ST R K++N + D+RG
Sbjct 573 TELEWDPSGRYVTTSSTIWRHKLENGYRLCDFRG 606
> At5g25780
Length=730
Score = 35.4 bits (80), Expect = 0.055, Method: Composition-based stats.
Identities = 31/132 (23%), Positives = 51/132 (38%), Gaps = 13/132 (9%)
Query 42 WSPTRDEFVLIEGKSPSDIVLL------DRQGRTQ--FVFPRRYRNFVSFCPFGQMAVVA 93
W P F +I G P V GR + N + + P G+ ++A
Sbjct 485 WEPKGHRFAVIHGDQPRPDVSFYSMKTAQNTGRVSKLATLKAKQANALFWSPTGKYIILA 544
Query 94 GFGNLAGEVSFYYRDKASQGMQHISEFKAPCTVLALWAPCGSYFATASTYPRMKVDNCLS 153
G G++ F+ D+ + M F A W P G Y AT+ T +++N +
Sbjct 545 GLKGFNGQLEFFNVDEL-ETMATAEHFMATDIE---WDPTGRYVATSVTTVH-EMENGFT 599
Query 154 VLDYRGESVQRL 165
+ + G V R+
Sbjct 600 IWSFNGNMVYRI 611
> At5g27640
Length=738
Score = 33.9 bits (76), Expect = 0.16, Method: Composition-based stats.
Identities = 31/132 (23%), Positives = 51/132 (38%), Gaps = 13/132 (9%)
Query 42 WSPTRDEFVLIEGKSPSDIVLL------DRQGRTQ--FVFPRRYRNFVSFCPFGQMAVVA 93
W P F +I G P V GR + N + + P G+ ++A
Sbjct 494 WEPKGHRFAVIHGDQPRPDVSFYSMKTAQNTGRVSKLATLKAKQANALFWSPTGKYIILA 553
Query 94 GFGNLAGEVSFYYRDKASQGMQHISEFKAPCTVLALWAPCGSYFATASTYPRMKVDNCLS 153
G G++ F+ D+ + M F A W P G Y ATA T +++N +
Sbjct 554 GLKGFNGQLEFFNVDEL-ETMATAEHFMATDIE---WDPTGRYVATAVTSVH-EMENGFT 608
Query 154 VLDYRGESVQRL 165
+ + G + R+
Sbjct 609 IWSFNGIMLYRI 620
> CE20310
Length=725
Score = 30.4 bits (67), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 25/64 (39%), Gaps = 2/64 (3%)
Query 79 NFVSFCPFGQMAVVAGFGNLAGEVSFYYRDKASQGMQHISEFKAPCTVLALWAPCGSYFA 138
N V F P G V + G V Y+ D + + + + P W P G YF
Sbjct 516 NEVQFAPKGGWLAVLAKVSAGGNV--YFIDTSLSEAKRTNVIEHPLFNKGYWDPTGRYFV 573
Query 139 TAST 142
T ST
Sbjct 574 TCST 577
> CE04226
Length=326
Score = 30.0 bits (66), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 30/65 (46%), Gaps = 2/65 (3%)
Query 34 KKIVQDVTWSPTRDEFVLIEGKSPSD--IVLLDRQGRTQFVFPRRYRNFVSFCPFGQMAV 91
++I++ W F+ G S S ++LL+ F FP RY N+V+ +AV
Sbjct 91 QEIIKPPQWINVAQHFLTWVGLSASSASLILLNADKLLYFKFPLRYANWVTSFKGVSLAV 150
Query 92 VAGFG 96
FG
Sbjct 151 FVWFG 155
> 7291366
Length=478
Score = 29.6 bits (65), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 34 KKIVQDVTWSPTRDEFVLIEGKSPSDIVLLDR 65
+K++QDV W PT++ + G PS+I +L R
Sbjct 403 RKLLQDVCWDPTQELSTCLRGFLPSNICVLRR 434
> At2g20330
Length=648
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 20/33 (60%), Gaps = 2/33 (6%)
Query 37 VQDVTWSPTRDEFVLIEGKSPSDIVLLDRQGRT 69
V+ V+WSPT +F+ + G + + I DR G T
Sbjct 227 VRSVSWSPTSGQFLCVTGSAQAKI--FDRDGLT 257
Lambda K H
0.325 0.136 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2425968562
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40