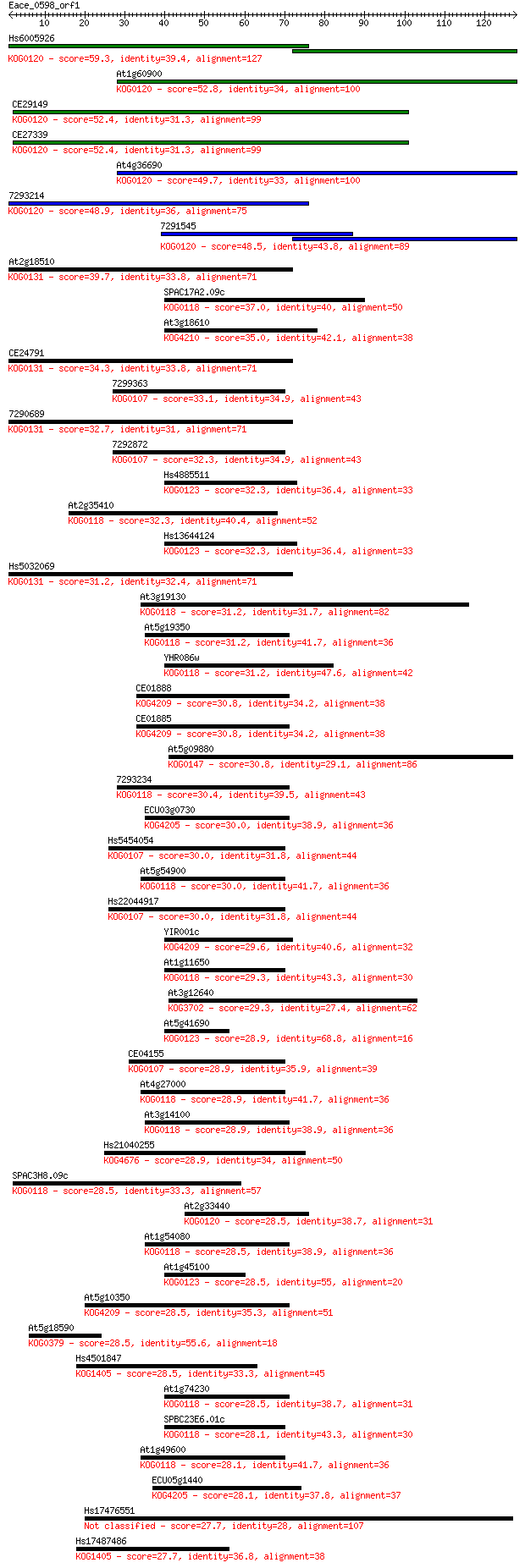
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0598_orf1
Length=127
Score E
Sequences producing significant alignments: (Bits) Value
Hs6005926 59.3 2e-09
At1g60900 52.8 2e-07
CE29149 52.4 2e-07
CE27339 52.4 2e-07
At4g36690 49.7 1e-06
7293214 48.9 2e-06
7291545 48.5 3e-06
At2g18510 39.7 0.001
SPAC17A2.09c 37.0 0.008
At3g18610 35.0 0.032
CE24791 34.3 0.051
7299363 33.1 0.13
7290689 32.7 0.17
7292872 32.3 0.19
Hs4885511 32.3 0.21
At2g35410 32.3 0.21
Hs13644124 32.3 0.22
Hs5032069 31.2 0.42
At3g19130 31.2 0.43
At5g19350 31.2 0.47
YHR086w 31.2 0.53
CE01888 30.8 0.63
CE01885 30.8 0.67
At5g09880 30.8 0.71
7293234 30.4 0.82
ECU03g0730 30.0 0.99
Hs5454054 30.0 1.1
At5g54900 30.0 1.1
Hs22044917 30.0 1.2
YIR001c 29.6 1.5
At1g11650 29.3 1.8
At3g12640 29.3 1.9
At5g41690 28.9 2.1
CE04155 28.9 2.2
At4g27000 28.9 2.2
At3g14100 28.9 2.2
Hs21040255 28.9 2.5
SPAC3H8.09c 28.5 2.8
At2g33440 28.5 3.0
At1g54080 28.5 3.0
At1g45100 28.5 3.1
At5g10350 28.5 3.2
At5g18590 28.5 3.2
Hs4501847 28.5 3.3
At1g74230 28.5 3.5
SPBC23E6.01c 28.1 3.6
At1g49600 28.1 4.2
ECU05g1440 28.1 4.5
Hs17476551 27.7 4.7
Hs17487486 27.7 5.0
> Hs6005926
Length=475
Score = 59.3 bits (142), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/75 (40%), Positives = 46/75 (61%), Gaps = 2/75 (2%)
Query 1 LTEQMFQEMVWEEMKIRGFCSNPNVNPVLYVWFAKDKGNYGFVEFTSVEETERALTMDGM 60
+TE+ + +M++ G P NPVL V +DK N+ F+EF SV+ET +A+ DG+
Sbjct 160 ITEEAMMDFFNAQMRLGGLTQAPG-NPVLAVQINQDK-NFAFLEFRSVDETTQAMAFDGI 217
Query 61 LCLGVPLKVSRPNDY 75
+ G LK+ RP+DY
Sbjct 218 IFQGQSLKIRRPHDY 232
Score = 33.5 bits (75), Expect = 0.096, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 27/62 (43%), Gaps = 6/62 (9%)
Query 72 PNDYTTTSSAQTHAMTVMGQQAAAILL------GGALGPQQLDAAAMNSTRKLRRLYFGN 125
P + + Q AM GQ A LL G A+ P + TR+ RRLY GN
Sbjct 96 PPGFEHITPMQYKAMQAAGQIPATALLPTMTPDGLAVTPTPVPVVGSQMTRQARRLYVGN 155
Query 126 LP 127
+P
Sbjct 156 IP 157
> At1g60900
Length=568
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 34/100 (34%), Positives = 51/100 (51%), Gaps = 9/100 (9%)
Query 28 VLYVWFAKDKGNYGFVEFTSVEETERALTMDGMLCLGVPLKVSRPNDYTTTSSAQTHAMT 87
V+ V+ +K + FVE SVEE A+ +DG++ GVP+KV RP DY + +A T
Sbjct 272 VVNVYINHEK-KFAFVEMRSVEEASNAMALDGIILEGVPVKVRRPTDYNPSLAA-----T 325
Query 88 VMGQQAAAILLGGALGPQQLDAAAMNSTRKLRRLYFGNLP 127
+ Q L GA+G L + + R++ G LP
Sbjct 326 LGPSQPNPNLNLGAVG---LSSGSTGGLEGPDRIFVGGLP 362
> CE29149
Length=474
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 31/104 (29%), Positives = 51/104 (49%), Gaps = 7/104 (6%)
Query 2 TEQMFQEMVWEEMKIRGFCSNPNVNPVLYVWFAKDKGNYGFVEFTSVEETERALTMDGML 61
E+ + ++M + G P NP+L DK N+ F+EF S++ET + DG+
Sbjct 174 NEEAMLDFFNQQMHLCGLAQAPG-NPILLCQINLDK-NFAFIEFRSIDETTAGMAFDGIN 231
Query 62 CLGVPLKVSRPNDYTTTS-----SAQTHAMTVMGQQAAAILLGG 100
+G LKV RP DY + +++ T++ A I +GG
Sbjct 232 FMGQQLKVRRPRDYQPSQNTFDMNSRMPVSTIVVDSANKIFIGG 275
> CE27339
Length=496
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 31/104 (29%), Positives = 51/104 (49%), Gaps = 7/104 (6%)
Query 2 TEQMFQEMVWEEMKIRGFCSNPNVNPVLYVWFAKDKGNYGFVEFTSVEETERALTMDGML 61
E+ + ++M + G P NP+L DK N+ F+EF S++ET + DG+
Sbjct 196 NEEAMLDFFNQQMHLCGLAQAPG-NPILLCQINLDK-NFAFIEFRSIDETTAGMAFDGIN 253
Query 62 CLGVPLKVSRPNDYTTTS-----SAQTHAMTVMGQQAAAILLGG 100
+G LKV RP DY + +++ T++ A I +GG
Sbjct 254 FMGQQLKVRRPRDYQPSQNTFDMNSRMPVSTIVVDSANKIFIGG 297
> At4g36690
Length=573
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/100 (33%), Positives = 51/100 (51%), Gaps = 9/100 (9%)
Query 28 VLYVWFAKDKGNYGFVEFTSVEETERALTMDGMLCLGVPLKVSRPNDYTTTSSAQTHAMT 87
V+ V+ +K + FVE SVEE A+++DG++ G P+KV RP+DY + +A T
Sbjct 277 VVNVYINHEK-KFAFVEMRSVEEASNAMSLDGIIFEGAPVKVRRPSDYNPSLAA-----T 330
Query 88 VMGQQAAAILLGGALGPQQLDAAAMNSTRKLRRLYFGNLP 127
+ Q + L A+G L A R++ G LP
Sbjct 331 LGPSQPSPHLNLAAVG---LTPGASGGLEGPDRIFVGGLP 367
> 7293214
Length=416
Score = 48.9 bits (115), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 42/75 (56%), Gaps = 2/75 (2%)
Query 1 LTEQMFQEMVWEEMKIRGFCSNPNVNPVLYVWFAKDKGNYGFVEFTSVEETERALTMDGM 60
+TE+ E ++M + G +PVL DK N+ F+EF S++ET +A+ DG+
Sbjct 104 VTEEEMMEFFNQQMHLVGLAQAAG-SPVLACQINLDK-NFAFLEFRSIDETTQAMAFDGI 161
Query 61 LCLGVPLKVSRPNDY 75
G LK+ RP+DY
Sbjct 162 NLKGQSLKIRRPHDY 176
> 7291545
Length=449
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 21/48 (43%), Positives = 29/48 (60%), Gaps = 0/48 (0%)
Query 39 NYGFVEFTSVEETERALTMDGMLCLGVPLKVSRPNDYTTTSSAQTHAM 86
N+ F+EF S++E +AL DGM+ G LK+ RP+DY S AM
Sbjct 163 NFAFLEFRSIDEASQALNFDGMVFRGQTLKIRRPHDYQPVPSISVSAM 210
Score = 37.0 bits (84), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 23/56 (41%), Positives = 28/56 (50%), Gaps = 2/56 (3%)
Query 72 PNDYTTTSSAQTHAMTVMGQQAAAILLGGALGPQQLDAAAMNSTRKLRRLYFGNLP 127
P Y + Q AM GQ A+ IL G G + AA TR+ RRLY GN+P
Sbjct 66 PEGYNHLTPQQYKAMLASGQIASRILSDGVHGGE--SAAISMITRQARRLYVGNIP 119
> At2g18510
Length=363
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 42/77 (54%), Gaps = 14/77 (18%)
Query 1 LTEQMFQEMVWEEMKIRGFCSNPNVNPVLYVWFAKDK-----GNYGFVEFTSVEETERAL 55
L Q+ +E++WE G PV+ V+ KD+ NYGF+E+ S E+ + A+
Sbjct 32 LDAQLSEELLWELFVQAG--------PVVNVYVPKDRVTNLHQNYGFIEYRSEEDADYAI 83
Query 56 TMDGMLCL-GVPLKVSR 71
+ M+ L G P++V++
Sbjct 84 KVLNMIKLHGKPIRVNK 100
> SPAC17A2.09c
Length=632
Score = 37.0 bits (84), Expect = 0.008, Method: Composition-based stats.
Identities = 20/51 (39%), Positives = 30/51 (58%), Gaps = 1/51 (1%)
Query 40 YGFVEFTSVEETERALT-MDGMLCLGVPLKVSRPNDYTTTSSAQTHAMTVM 89
YGFV F+S +E + AL M G LC G PL++S + + S A A+ ++
Sbjct 226 YGFVRFSSEKEQQHALMHMQGYLCQGRPLRISVASPKSRASIAADSALGIV 276
> At3g18610
Length=636
Score = 35.0 bits (79), Expect = 0.032, Method: Composition-based stats.
Identities = 16/38 (42%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 40 YGFVEFTSVEETERALTMDGMLCLGVPLKVSRPNDYTT 77
YG +EF S EE ++AL M+G L LG +++ N+ T
Sbjct 426 YGHIEFASPEEAQKALEMNGKLLLGRDVRLDLANERGT 463
> CE24791
Length=389
Score = 34.3 bits (77), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 40/77 (51%), Gaps = 14/77 (18%)
Query 1 LTEQMFQEMVWEEMKIRGFCSNPNVNPVLYVWFAKDK-----GNYGFVEFTSVEETERAL 55
L E++ + ++WE M G PV+ V KD+ +GFVEF E+ + A+
Sbjct 30 LDEKVSESILWELMVQAG--------PVVSVNMPKDRVTANHQGFGFVEFMGEEDADYAI 81
Query 56 TMDGMLCL-GVPLKVSR 71
+ M+ L G P+KV++
Sbjct 82 KILNMIKLYGKPIKVNK 98
> 7299363
Length=135
Score = 33.1 bits (74), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 23/44 (52%), Gaps = 1/44 (2%)
Query 27 PVLYVWFAKDKGNYGFVEFTSVEETERAL-TMDGMLCLGVPLKV 69
P+ VW A++ + FVEF + E A +DG C G ++V
Sbjct 36 PLRNVWVARNPPGFAFVEFEDRRDAEDATRALDGTRCCGTRIRV 79
> 7290689
Length=347
Score = 32.7 bits (73), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 22/77 (28%), Positives = 39/77 (50%), Gaps = 14/77 (18%)
Query 1 LTEQMFQEMVWEEMKIRGFCSNPNVNPVLYVWFAKDK-----GNYGFVEFTSVEETERAL 55
L +++ + ++WE G PV+ V KD+ YGFVEF S E+ + +
Sbjct 20 LDDKVSETLLWELFVQAG--------PVVNVHMPKDRVTQMHQGYGFVEFLSEEDADYGI 71
Query 56 TMDGMLCL-GVPLKVSR 71
+ M+ L G P++V++
Sbjct 72 KIMNMIKLYGKPIRVNK 88
> 7292872
Length=130
Score = 32.3 bits (72), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 23/44 (52%), Gaps = 1/44 (2%)
Query 27 PVLYVWFAKDKGNYGFVEFTSVEETERALT-MDGMLCLGVPLKV 69
P+ VW A++ + FVEF + E A +DG C G ++V
Sbjct 36 PLRNVWVARNPPGFAFVEFEDRRDAEDATRGLDGTRCCGTRIRV 79
> Hs4885511
Length=707
Score = 32.3 bits (72), Expect = 0.21, Method: Composition-based stats.
Identities = 12/33 (36%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 40 YGFVEFTSVEETERALTMDGMLCLGVPLKVSRP 72
+G+V+F S E+ E+AL + G+ G +K+ +P
Sbjct 349 FGYVDFESAEDLEKALELTGLKVFGNEIKLEKP 381
> At2g35410
Length=308
Score = 32.3 bits (72), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 31/57 (54%), Gaps = 5/57 (8%)
Query 16 IRGFCSNPNVNPV-LYVWFAKDKGN---YGFVEFTSVEETERALT-MDGMLCLGVPL 67
+R + + NPV V FA +G YGFV F + EE E A+T ++G +G P+
Sbjct 210 LRELFTAADFNPVSARVVFADPEGRSSGYGFVSFATREEAENAITKLNGKEIMGRPI 266
> Hs13644124
Length=710
Score = 32.3 bits (72), Expect = 0.22, Method: Composition-based stats.
Identities = 12/33 (36%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 40 YGFVEFTSVEETERALTMDGMLCLGVPLKVSRP 72
+G+V+F S E+ E+AL + G+ G +K+ +P
Sbjct 349 FGYVDFESAEDLEKALELTGLKVFGNEIKLEKP 381
> Hs5032069
Length=424
Score = 31.2 bits (69), Expect = 0.42, Method: Composition-based stats.
Identities = 23/77 (29%), Positives = 39/77 (50%), Gaps = 14/77 (18%)
Query 1 LTEQMFQEMVWEEMKIRGFCSNPNVNPVLYVWFAKDK-----GNYGFVEFTSVEETERAL 55
L E++ + ++WE G PV+ KD+ YGFVEF S E+ + A+
Sbjct 20 LDEKVSEPLLWELFLQAG--------PVVNTHMPKDRVTGQHQGYGFVEFLSEEDADYAI 71
Query 56 TMDGMLCL-GVPLKVSR 71
+ M+ L G P++V++
Sbjct 72 KIMNMIKLYGKPIRVNK 88
> At3g19130
Length=435
Score = 31.2 bits (69), Expect = 0.43, Method: Composition-based stats.
Identities = 26/90 (28%), Positives = 38/90 (42%), Gaps = 16/90 (17%)
Query 34 AKDKGNYGFVEFTSVEETERALT-MDGMLCLGVPLKVSRPNDYTTTSSAQTHAMTVMGQQ 92
+ KG YGFV F E RALT M+G C ++V ++ Q H+
Sbjct 241 GRSKG-YGFVRFGDENERSRALTEMNGAYCSNRQMRVGIATPKRAIANQQQHS------- 292
Query 93 AAAILLGGALGPQ-------QLDAAAMNST 115
+ A++L G G Q D + N+T
Sbjct 293 SQAVILAGGHGSNGSMGYGSQSDGESTNAT 322
> At5g19350
Length=425
Score = 31.2 bits (69), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 23/37 (62%), Gaps = 2/37 (5%)
Query 35 KDKGNYGFVEFTSVEETERALT-MDGMLCLGVPLKVS 70
+ KG YGFV+F E RA+ M+G+ C P+++S
Sbjct 156 RSKG-YGFVKFAEESERNRAMAEMNGLYCSTRPMRIS 191
> YHR086w
Length=523
Score = 31.2 bits (69), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 29/53 (54%), Gaps = 11/53 (20%)
Query 40 YGFVEFTSVEETERALT-MDGMLCLGVPLK----------VSRPNDYTTTSSA 81
YGFV+FT+ +E + AL+ M G+ G +K VS NDY +SS+
Sbjct 207 YGFVKFTNSDEQQLALSEMQGVFLNGRAIKVGPTSGQQQHVSGNNDYNRSSSS 259
> CE01888
Length=237
Score = 30.8 bits (68), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 33 FAKDKGNYGFVEFTSVEETERALTMDGMLCLGVPLKVS 70
F K + N+ ++EF E AL M+G L P+ V+
Sbjct 137 FTKKQKNFAYIEFDDSSSIENALVMNGSLFRSRPIVVT 174
> CE01885
Length=197
Score = 30.8 bits (68), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 33 FAKDKGNYGFVEFTSVEETERALTMDGMLCLGVPLKVS 70
F K + N+ ++EF E AL M+G L P+ V+
Sbjct 97 FTKKQKNFAYIEFDDSSSIENALVMNGSLFRSRPIVVT 134
> At5g09880
Length=604
Score = 30.8 bits (68), Expect = 0.71, Method: Composition-based stats.
Identities = 25/86 (29%), Positives = 38/86 (44%), Gaps = 25/86 (29%)
Query 41 GFVEFTSVEETERALTMDGMLCLGVPLKVSRPNDYTTTSSAQTHAMTVMGQQAAAILLGG 100
G++EF V A+ + G L LG P+ V +P++ + AQ+++ TV G
Sbjct 212 GYIEFYDVMSVPMAIALSGQLFLGQPVMV-KPSE-AEKNLAQSNSTTV-----------G 258
Query 101 ALGPQQLDAAAMNSTRKLRRLYFGNL 126
GP R+LY GNL
Sbjct 259 GTGPAD------------RKLYVGNL 272
> 7293234
Length=325
Score = 30.4 bits (67), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 8/51 (15%)
Query 28 VLYVWFAKDK-----GNYGFVEFTSVEETERALTMDGML---CLGVPLKVS 70
V YV KD+ + +VEF +++ ERAL DG + L PL++
Sbjct 54 VKYVRLVKDRETDQFKGFCYVEFETLDNLERALECDGRIKLDDLSAPLRID 104
> ECU03g0730
Length=293
Score = 30.0 bits (66), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 25/36 (69%), Gaps = 1/36 (2%)
Query 35 KDKGNYGFVEFTSVEETERALTMDGMLCLGVPLKVS 70
++KG +G+VEF E+ ++AL +DG + LG + V+
Sbjct 126 RNKG-FGYVEFCKEEDVKKALKLDGTVFLGREVVVN 160
> Hs5454054
Length=238
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 24/45 (53%), Gaps = 1/45 (2%)
Query 26 NPVLYVWFAKDKGNYGFVEFTSVEETERALT-MDGMLCLGVPLKV 69
P+ VW A++ + FVEF + E A+ +DG + G ++V
Sbjct 35 GPLRTVWIARNPPGFAFVEFEDPRDAEDAVRGLDGKVICGSRVRV 79
> At5g54900
Length=390
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 21/37 (56%), Gaps = 2/37 (5%)
Query 34 AKDKGNYGFVEFTSVEETERALT-MDGMLCLGVPLKV 69
+ KG YGFV F E RA+T M+G C P+++
Sbjct 193 GRSKG-YGFVRFADENEQMRAMTEMNGQYCSTRPMRI 228
> Hs22044917
Length=137
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 24/45 (53%), Gaps = 1/45 (2%)
Query 26 NPVLYVWFAKDKGNYGFVEFTSVEETERALT-MDGMLCLGVPLKV 69
P+ VW A++ + FVEF + E A+ +DG + G ++V
Sbjct 35 GPLRTVWIARNPPGFAFVEFEDPRDAEDAVRGLDGKVICGSRVRV 79
> YIR001c
Length=250
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 40 YGFVEFTSVEETERALTMDGMLCLGVPLKVSR 71
YG++EF S E+AL ++G G + VSR
Sbjct 107 YGYIEFESPAYREKALQLNGGELKGKKIAVSR 138
> At1g11650
Length=405
Score = 29.3 bits (64), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 20/31 (64%), Gaps = 1/31 (3%)
Query 40 YGFVEFTSVEETERALT-MDGMLCLGVPLKV 69
YGFV F+ E RA+T M+G+ C P+++
Sbjct 199 YGFVRFSDESEQIRAMTEMNGVPCSTRPMRI 229
> At3g12640
Length=674
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 17/64 (26%), Positives = 34/64 (53%), Gaps = 2/64 (3%)
Query 41 GFVEFTSVEETERALTMDGMLCLGVPLKVSRPNDYTTTSSAQTHAMTVMGQ--QAAAILL 98
++EFT E E AL++DG + LK+ + ++ +A + + + G+ +A +
Sbjct 560 AYIEFTRKEAAENALSLDGTSFMSRILKIVKGSNGQNQEAASSMSWSRGGRFTRAPSYFR 619
Query 99 GGAL 102
GGA+
Sbjct 620 GGAV 623
> At5g41690
Length=599
Score = 28.9 bits (63), Expect = 2.1, Method: Composition-based stats.
Identities = 11/16 (68%), Positives = 14/16 (87%), Gaps = 0/16 (0%)
Query 40 YGFVEFTSVEETERAL 55
YGFVEF S +ET++AL
Sbjct 214 YGFVEFASADETKKAL 229
> CE04155
Length=179
Score = 28.9 bits (63), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 21/40 (52%), Gaps = 1/40 (2%)
Query 31 VWFAKDKGNYGFVEFTSVEETERAL-TMDGMLCLGVPLKV 69
VW A+ + FVE+ V + E A+ +DG GV +V
Sbjct 32 VWVARRPPGFAFVEYDDVRDAEDAVRALDGSRICGVRARV 71
> At4g27000
Length=427
Score = 28.9 bits (63), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 20/37 (54%), Gaps = 2/37 (5%)
Query 34 AKDKGNYGFVEFTSVEETERALT-MDGMLCLGVPLKV 69
+ KG YGFV F E RA+T M+G C P++
Sbjct 212 GRSKG-YGFVRFADESEQIRAMTEMNGQYCSSRPMRT 247
> At3g14100
Length=427
Score = 28.9 bits (63), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 21/37 (56%), Gaps = 1/37 (2%)
Query 35 KDKGNYGFVEFTSVEETERA-LTMDGMLCLGVPLKVS 70
KDK +YGFV + A L+++G G P+KV+
Sbjct 93 KDKSSYGFVHYFDRRSAALAILSLNGRHLFGQPIKVN 129
> Hs21040255
Length=508
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 26/53 (49%), Gaps = 3/53 (5%)
Query 25 VNPVLYVWFAKDKGN---YGFVEFTSVEETERALTMDGMLCLGVPLKVSRPND 74
V V +V A D+ + FVEF RAL +G++ PLK++ N+
Sbjct 90 VGEVKFVRMAGDETQPTRFAFVEFADQNSVPRALAFNGVMFGDRPLKINHSNN 142
> SPAC3H8.09c
Length=738
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 19/57 (33%), Positives = 28/57 (49%), Gaps = 11/57 (19%)
Query 2 TEQMFQEMVWEEMKIRGFCSNPNVNPVLYVWFAKDKGNYGFVEFTSVEETERALTMD 58
T+ + + +W+ KI G P VL K NYGFV+F + E+ RAL +
Sbjct 372 TKSLSKRNLWKVFKIYG----PLAQIVL-------KANYGFVQFFTNEDCARALNAE 417
> At2g33440
Length=475
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 12/31 (38%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 45 FTSVEETERALTMDGMLCLGVPLKVSRPNDY 75
F + ++ AL++DG G LK+ RP DY
Sbjct 253 FLTPQDASAALSLDGCSFAGSNLKIRRPKDY 283
> At1g54080
Length=426
Score = 28.5 bits (62), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 21/37 (56%), Gaps = 1/37 (2%)
Query 35 KDKGNYGFVEFTSVEETERAL-TMDGMLCLGVPLKVS 70
KDK +YGFV + A+ T++G G P+KV+
Sbjct 97 KDKSSYGFVHYFDRRCASMAIMTLNGRHIFGQPMKVN 133
> At1g45100
Length=497
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 11/20 (55%), Positives = 14/20 (70%), Gaps = 0/20 (0%)
Query 40 YGFVEFTSVEETERALTMDG 59
+GFVEF S E E+AL +G
Sbjct 289 WGFVEFASANEAEKALVKNG 308
> At5g10350
Length=217
Score = 28.5 bits (62), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 25/51 (49%), Gaps = 1/51 (1%)
Query 20 CSNPNVNPVLYVWFAKDKGNYGFVEFTSVEETERALTMDGMLCLGVPLKVS 70
C N +L F + KG + +VEF VE + AL ++ G LKVS
Sbjct 112 CGTVNRVTILMDKFGQPKG-FAYVEFVEVEAVQEALQLNESELHGRQLKVS 161
> At5g18590
Length=792
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 10/18 (55%), Positives = 13/18 (72%), Gaps = 0/18 (0%)
Query 6 FQEMVWEEMKIRGFCSNP 23
F+ MVW +KIRGF +P
Sbjct 334 FETMVWSRIKIRGFHPSP 351
> Hs4501847
Length=500
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 23/46 (50%), Gaps = 5/46 (10%)
Query 18 GFCSNPNVNPVLYVWF-AKDKGNYGFVEFTSVEETERALTMDGMLC 62
G CSN N +++W+ +K++G GF S EE E + C
Sbjct 164 GSCSNENALKTIFMWYRSKERGQRGF----SKEELETCMINQAPWC 205
> At1g74230
Length=289
Score = 28.5 bits (62), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 40 YGFVEFTSVEETERALTMDGMLCLGVPLKVS 70
+ FV FTS EE A+ +DG G ++V+
Sbjct 77 FAFVTFTSTEEASNAMQLDGQDLHGRRIRVN 107
> SPBC23E6.01c
Length=255
Score = 28.1 bits (61), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 18/31 (58%), Gaps = 1/31 (3%)
Query 40 YGFVEFTSVEETERALT-MDGMLCLGVPLKV 69
YGFV FT + + AL M G +C P++V
Sbjct 12 YGFVRFTDENDQKSALAEMQGQICGDRPIRV 42
> At1g49600
Length=445
Score = 28.1 bits (61), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 20/37 (54%), Gaps = 2/37 (5%)
Query 34 AKDKGNYGFVEFTSVEETERALT-MDGMLCLGVPLKV 69
+ KG YGFV F E RA+T M+G C ++V
Sbjct 252 GRSKG-YGFVRFGDENERSRAMTEMNGAFCSSRQMRV 287
> ECU05g1440
Length=301
Score = 28.1 bits (61), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 16/37 (43%), Gaps = 0/37 (0%)
Query 37 KGNYGFVEFTSVEETERALTMDGMLCLGVPLKVSRPN 73
KG YGF+ F S R L G P+ V R N
Sbjct 33 KGTYGFLNFDSEGSITRVLNQRTHSIDGAPISVERAN 69
> Hs17476551
Length=293
Score = 27.7 bits (60), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 30/121 (24%), Positives = 46/121 (38%), Gaps = 17/121 (14%)
Query 20 CSNPNVNPVLYVWFAKDKGNYGFVEFTSVEETERALTMDGMLCLGVPLKVSRPNDYTTTS 79
C+ N +P W+ +D G G V + E E+ + + L + T
Sbjct 43 CNYTNYSPAYLQWYRQDPGR-GPVFLLLIRENEKEKRKERLKTHSSALAAASCTQTLLTL 101
Query 80 SAQTHAMTVMGQQ-AAAILLGG-------------ALGPQQLDAAAMNSTRKLRRLYFGN 125
+ + H V + A +GG LGPQQ D A+M+ T + R F N
Sbjct 102 TIRAHGCAVSQRLITAPDRMGGWANGENQVEHSPHFLGPQQGDVASMSCTYSVSR--FNN 159
Query 126 L 126
L
Sbjct 160 L 160
> Hs17487486
Length=500
Score = 27.7 bits (60), Expect = 5.0, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 22/39 (56%), Gaps = 5/39 (12%)
Query 18 GFCSNPNVNPVLYVWF-AKDKGNYGFVEFTSVEETERAL 55
G CSN N +++W+ +K++G GF S EE E +
Sbjct 164 GSCSNENALKTIFMWYRSKERGQRGF----SQEELETCM 198
Lambda K H
0.319 0.133 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1176738752
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40