bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0579_orf4
Length=744
Score E
Sequences producing significant alignments: (Bits) Value
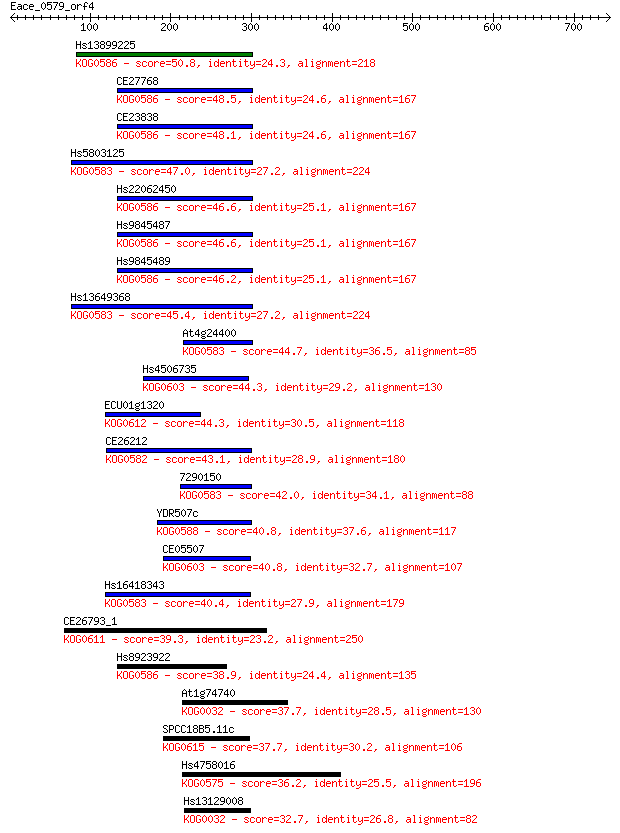
Hs13899225 50.8 1e-05
CE27768 48.5 6e-05
CE23838 48.1 6e-05
Hs5803125 47.0 2e-04
Hs22062450 46.6 2e-04
Hs9845487 46.6 2e-04
Hs9845489 46.2 3e-04
Hs13649368 45.4 4e-04
At4g24400 44.7 8e-04
Hs4506735 44.3 0.001
ECU01g1320 44.3 0.001
CE26212 43.1 0.002
7290150 42.0 0.005
YDR507c 40.8 0.010
CE05507 40.8 0.011
Hs16418343 40.4 0.014
CE26793_1 39.3 0.031
Hs8923922 38.9 0.048
At1g74740 37.7 0.085
SPCC18B5.11c 37.7 0.093
Hs4758016 36.2 0.25
Hs13129008 32.7 3.4
> Hs13899225
Length=688
Score = 50.8 bits (120), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 53/226 (23%), Positives = 93/226 (41%), Gaps = 33/226 (14%)
Query 83 VAKIKGMPDVLHTQHFTVLSTVEKMENIQVTLRDVLDVLDWVPTGTRA------FLARHV 136
V +KG+ + F V+ T EK + + +V D++ + R R +
Sbjct 107 VRIMKGLNHPNIVKLFEVIET-EKTLYLVMEYASAGEVFDYLVSHGRMKEKEARAKFRQI 165
Query 137 LKTVLHLQWAGWSNNQINSSSFGVQDDGSVLL--FGFESSVPFGSVIPADADVSAVYTEP 194
+ V + + + + + + + ++ + FGF + GS + S Y P
Sbjct 166 VSAVHYCHQKNIVHRDLKAENLLLDAEANIKIADFGFSNEFTLGSKLDTFCG-SPPYAAP 224
Query 195 ELLASIVCEESCEFQPIANPKADLWSLGVLLYEILMDGELPFGLSSLPPDEETVAYVVQL 254
EL + + P+ D+WSLGV+LY L+ G LPF +L E V
Sbjct 225 ELF---------QGKKYDGPEVDIWSLGVILY-TLVSGSLPFDGHNLKELRERVLR---- 270
Query 255 GAHASPESLTVDMEYRGIHKRWQSLIVRLLNPDREKRISAEEIVKD 300
G + P ++ D E S++ R L + KR + E+I+KD
Sbjct 271 GKYRVPFYMSTDCE---------SILRRFLVLNPAKRCTLEQIMKD 307
> CE27768
Length=1096
Score = 48.5 bits (114), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 41/169 (24%), Positives = 74/169 (43%), Gaps = 26/169 (15%)
Query 134 RHVLKTVLHLQWAGWSNNQINSSSFGVQDDGSVLL--FGFESSVPFGSVIPADADVSAVY 191
R ++ V +L + + + + + D ++ + FGF ++ G+ + S Y
Sbjct 226 RQIVSAVQYLHSKNIIHRDLKAENLLLDQDMNIKIADFGFSNTFSLGNKLDTFCG-SPPY 284
Query 192 TEPELLASIVCEESCEFQPIANPKADLWSLGVLLYEILMDGELPFGLSSLPPDEETVAYV 251
PEL + + P+ D+WSLGV+LY L+ G LPF +L E V
Sbjct 285 AAPELFSG---------KKYDGPEVDVWSLGVILY-TLVSGSLPFDGQNLKELRERVLR- 333
Query 252 VQLGAHASPESLTVDMEYRGIHKRWQSLIVRLLNPDREKRISAEEIVKD 300
G + P ++ D E +L+ + L + ++R S + I+KD
Sbjct 334 ---GKYRIPFYMSTDCE---------NLLKKFLVINPQRRSSLDNIMKD 370
> CE23838
Length=1192
Score = 48.1 bits (113), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 41/169 (24%), Positives = 74/169 (43%), Gaps = 26/169 (15%)
Query 134 RHVLKTVLHLQWAGWSNNQINSSSFGVQDDGSVLL--FGFESSVPFGSVIPADADVSAVY 191
R ++ V +L + + + + + D ++ + FGF ++ G+ + S Y
Sbjct 274 RQIVSAVQYLHSKNIIHRDLKAENLLLDQDMNIKIADFGFSNTFSLGNKLDTFCG-SPPY 332
Query 192 TEPELLASIVCEESCEFQPIANPKADLWSLGVLLYEILMDGELPFGLSSLPPDEETVAYV 251
PEL + + P+ D+WSLGV+LY L+ G LPF +L E V
Sbjct 333 AAPELFSG---------KKYDGPEVDVWSLGVILY-TLVSGSLPFDGQNLKELRERVLR- 381
Query 252 VQLGAHASPESLTVDMEYRGIHKRWQSLIVRLLNPDREKRISAEEIVKD 300
G + P ++ D E +L+ + L + ++R S + I+KD
Sbjct 382 ---GKYRIPFYMSTDCE---------NLLKKFLVINPQRRSSLDNIMKD 418
> Hs5803125
Length=334
Score = 47.0 bits (110), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 61/235 (25%), Positives = 93/235 (39%), Gaps = 46/235 (19%)
Query 77 IAAITSVAKIKGMPDVLHTQHFTVLSTVEKMENIQVTLRDVL---DVLDWV----PTG-- 127
+A + V G P V+ +L E E + L L D+ D++ P G
Sbjct 84 VALLWKVGAGGGHPGVIR-----LLDWFETQEGFMLVLERPLPAQDLFDYITEKGPLGEG 138
Query 128 -TRAFLARHVLKTVLHLQWAGWSNNQINSSSFGVQ-DDGSVLLFGFESSVPFGSVIPADA 185
+R F + V+ + H G + I + + G L F S D
Sbjct 139 PSRCFFGQ-VVAAIQHCHSRGVVHRDIKDENILIDLRRGCAKLIDFGSGALLHDEPYTDF 197
Query 186 DVSAVYTEPELLASIVCEESCEFQPIANPKADLWSLGVLLYEILMDGELPFGLSSLPPDE 245
D + VY+ PE ++ Q A P A +WSLG+LLY+++ G++PF D+
Sbjct 198 DGTRVYSPPEWIS--------RHQYHALP-ATVWSLGILLYDMVC-GDIPF-----ERDQ 242
Query 246 ETVAYVVQLGAHASPESLTVDMEYRGIHKRWQSLIVRLLNPDREKRISAEEIVKD 300
E + + AH SP+ +LI R L P R S EEI+ D
Sbjct 243 EILEAELHFPAHVSPDCC--------------ALIRRCLAPKPSSRPSLEEILLD 283
> Hs22062450
Length=787
Score = 46.6 bits (109), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 42/169 (24%), Positives = 72/169 (42%), Gaps = 26/169 (15%)
Query 134 RHVLKTVLHLQWAGWSNNQINSSSFGVQDDGSVLL--FGFESSVPFGSVIPADADVSAVY 191
R ++ V + + + + + + D ++ + FGF + FG+ + S Y
Sbjct 157 RQIVSAVQYCHQKFIVHRDLKAENLLLDADMNIKIADFGFSNEFTFGNKLDTFCG-SPPY 215
Query 192 TEPELLASIVCEESCEFQPIANPKADLWSLGVLLYEILMDGELPFGLSSLPPDEETVAYV 251
PEL + P+ D+WSLGV+LY L+ G LPF +L E V
Sbjct 216 AAPELFQG---------KKYDGPEVDVWSLGVILY-TLVSGSLPFDGQNLKELRERVLR- 264
Query 252 VQLGAHASPESLTVDMEYRGIHKRWQSLIVRLLNPDREKRISAEEIVKD 300
G + P ++ D E +L+ + L + KR + E+I+KD
Sbjct 265 ---GKYRIPFYMSTDCE---------NLLKKFLILNPSKRGTLEQIMKD 301
> Hs9845487
Length=745
Score = 46.6 bits (109), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 42/169 (24%), Positives = 72/169 (42%), Gaps = 26/169 (15%)
Query 134 RHVLKTVLHLQWAGWSNNQINSSSFGVQDDGSVLL--FGFESSVPFGSVIPADADVSAVY 191
R ++ V + + + + + + D ++ + FGF + FG+ + S Y
Sbjct 124 RQIVSAVQYCHQKFIVHRDLKAENLLLDADMNIKIADFGFSNEFTFGNKLDTFCG-SPPY 182
Query 192 TEPELLASIVCEESCEFQPIANPKADLWSLGVLLYEILMDGELPFGLSSLPPDEETVAYV 251
PEL + P+ D+WSLGV+LY L+ G LPF +L E V
Sbjct 183 AAPELFQG---------KKYDGPEVDVWSLGVILY-TLVSGSLPFDGQNLKELRERVLR- 231
Query 252 VQLGAHASPESLTVDMEYRGIHKRWQSLIVRLLNPDREKRISAEEIVKD 300
G + P ++ D E +L+ + L + KR + E+I+KD
Sbjct 232 ---GKYRIPFYMSTDCE---------NLLKKFLILNPSKRGTLEQIMKD 268
> Hs9845489
Length=691
Score = 46.2 bits (108), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 42/169 (24%), Positives = 72/169 (42%), Gaps = 26/169 (15%)
Query 134 RHVLKTVLHLQWAGWSNNQINSSSFGVQDDGSVLL--FGFESSVPFGSVIPADADVSAVY 191
R ++ V + + + + + + D ++ + FGF + FG+ + S Y
Sbjct 124 RQIVSAVQYCHQKFIVHRDLKAENLLLDADMNIKIADFGFSNEFTFGNKLDTFCG-SPPY 182
Query 192 TEPELLASIVCEESCEFQPIANPKADLWSLGVLLYEILMDGELPFGLSSLPPDEETVAYV 251
PEL + P+ D+WSLGV+LY L+ G LPF +L E V
Sbjct 183 AAPELFQG---------KKYDGPEVDVWSLGVILY-TLVSGSLPFDGQNLKELRERVLR- 231
Query 252 VQLGAHASPESLTVDMEYRGIHKRWQSLIVRLLNPDREKRISAEEIVKD 300
G + P ++ D E +L+ + L + KR + E+I+KD
Sbjct 232 ---GKYRIPFYMSTDCE---------NLLKKFLILNPSKRGTLEQIMKD 268
> Hs13649368
Length=311
Score = 45.4 bits (106), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 61/235 (25%), Positives = 93/235 (39%), Gaps = 46/235 (19%)
Query 77 IAAITSVAKIKGMPDVLHTQHFTVLSTVEKMENIQVTLRDVL---DVLDWV----PTG-- 127
+A + V G P V+ +L E E + L L D+ D++ P G
Sbjct 84 VALLWKVGAGGGHPGVIR-----LLDWFETQEGFMLVLERPLPAQDLFDYITEKGPLGEG 138
Query 128 -TRAFLARHVLKTVLHLQWAGWSNNQINSSSFGVQ-DDGSVLLFGFESSVPFGSVIPADA 185
+R F + V+ + H G + I + + G L F S D
Sbjct 139 PSRCFFGQ-VVAAIQHCHSRGVVHRDIKDENILIDLRRGCAKLIDFGSGALLHDEPYTDF 197
Query 186 DVSAVYTEPELLASIVCEESCEFQPIANPKADLWSLGVLLYEILMDGELPFGLSSLPPDE 245
D + VY+ PE ++ Q A P A +WSLG+LLY+++ G++PF D+
Sbjct 198 DGTRVYSPPEWIS--------RHQYHALP-ATVWSLGILLYDMVC-GDIPF-----ERDQ 242
Query 246 ETVAYVVQLGAHASPESLTVDMEYRGIHKRWQSLIVRLLNPDREKRISAEEIVKD 300
E + + AH SP+ +LI R L P R S EEI+ D
Sbjct 243 EILEAELHFPAHVSPDCC--------------ALIRRCLAPKPSSRPSLEEILLD 283
> At4g24400
Length=445
Score = 44.7 bits (104), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 43/85 (50%), Gaps = 14/85 (16%)
Query 216 ADLWSLGVLLYEILMDGELPFGLSSLPPDEETVAYVVQLGAHASPESLTVDMEYRGIHKR 275
AD+WS GV+LY +LM G LPF LP T+ + + P +
Sbjct 189 ADIWSCGVILY-VLMAGYLPFDEMDLP----TLYSKIDKAEFSCPSYFALGA-------- 235
Query 276 WQSLIVRLLNPDREKRISAEEIVKD 300
+SLI R+L+P+ E RI+ EI KD
Sbjct 236 -KSLINRILDPNPETRITIAEIRKD 259
> Hs4506735
Length=772
Score = 44.3 bits (103), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 38/132 (28%), Positives = 62/132 (46%), Gaps = 16/132 (12%)
Query 166 VLLFGFESSVPFGSVIPADADVSAV-YTEPELLASIVCEESCEFQPIANPKADLWSLGVL 224
++ FGF P +P + Y PELLA +ESC DLWSLGV+
Sbjct 549 IIDFGFARLRPQSPGVPMQTPCFTLQYAAPELLAQQGYDESC----------DLWSLGVI 598
Query 225 LYEILMDGELPFGLSSLPPDEETVAYVVQLGAHASPESLTVDME-YRGIHKRWQSLIVRL 283
LY +++ G++PF +S + A ++ ++D E ++G+ + + L+ L
Sbjct 599 LY-MMLSGQVPFQGASGQGGQSQAAEIM---CKIREGRFSLDGEAWQGVSEEAKELVRGL 654
Query 284 LNPDREKRISAE 295
L D KR+ E
Sbjct 655 LTVDPAKRLKLE 666
> ECU01g1320
Length=865
Score = 44.3 bits (103), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 36/126 (28%), Positives = 54/126 (42%), Gaps = 24/126 (19%)
Query 119 DVL--DWVPTGTRAFLARHVLKTVLHLQWAGWSNNQINSSSFGVQDDGSVLLFGFESSVP 176
DVL DWV F A ++ + L GW + + + + DG V L F S +
Sbjct 136 DVLEEDWVR-----FYAAEIVAALDELHKLGWIHRDLKPDNVLIGIDGHVKLADFGSCIK 190
Query 177 F------GSVIPADADVSAVYTEPELLASIVCEESCEFQPIANPKADLWSLGVLLYEILM 230
S+ D Y P++L S+ E CE+ D W+LGV++YE++
Sbjct 191 MKDGKARSSITVGTPD----YVSPDVLCSV--NEECEY----GEDVDFWTLGVIIYEMIY 240
Query 231 DGELPF 236
G PF
Sbjct 241 -GTTPF 245
> CE26212
Length=545
Score = 43.1 bits (100), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 52/195 (26%), Positives = 85/195 (43%), Gaps = 38/195 (19%)
Query 120 VLDWVPTGTRAFLARHVLKTVLHLQWAGWSNNQINSSSFGVQDDGSVLLFGFESSVPFGS 179
VLD V T + R VLK + + G + I + + + DDG++ + F S G
Sbjct 160 VLDEVSIAT---VLREVLKGLEYFHLNGQIHRDIKAGNILLADDGTIQIADFGVS---GW 213
Query 180 VIPADADVSA-----------VYTEPELLASIVCEESCEFQPIANPKADLWSLGVLLYEI 228
+ + D+S + PE++ + + +F KAD+WSLG+L E
Sbjct 214 LASSGGDLSRQKVRHTFVGTPCWMAPEVMEQV---QGYDF------KADIWSLGILAIE- 263
Query 229 LMDGELPFGLSSLPPDEETVAYVVQLGAHASPESLTVDME----YRGIHKRWQSLIVRLL 284
L G P+ PP + V+ L P +L + E Y+ K +++LI L
Sbjct 264 LATGTAPY--HKYPPMK-----VLMLTLQNDPPTLETNAERKDQYKAYGKSFKTLIRDCL 316
Query 285 NPDREKRISAEEIVK 299
D KR +A E++K
Sbjct 317 QKDPAKRPTASELLK 331
> 7290150
Length=582
Score = 42.0 bits (97), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 30/88 (34%), Positives = 44/88 (50%), Gaps = 14/88 (15%)
Query 212 ANPKADLWSLGVLLYEILMDGELPFGLSSLPPDEETVAYVVQLGAHASPESLTVDMEYRG 271
A P+ D+WS GV+LY +L G LPF +P T+ ++ G PE L
Sbjct 203 AGPEVDIWSCGVILYALLC-GTLPFDDEHVP----TLFRKIKSGIFPIPEYLN------- 250
Query 272 IHKRWQSLIVRLLNPDREKRISAEEIVK 299
K+ +L+ ++L D KR + EEI K
Sbjct 251 --KQVVNLVCQMLQVDPLKRANIEEIKK 276
> YDR507c
Length=1142
Score = 40.8 bits (94), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 44/133 (33%), Positives = 61/133 (45%), Gaps = 36/133 (27%)
Query 183 ADADVSAVYTEPELLASIVCEESCEFQPIANPK-----------ADLWSLGVLLYEILMD 231
AD ++A+ TE +LL E SC A P+ +D+WS GV+L+ +L
Sbjct 173 ADFGMAALETEGKLL-----ETSCGSPHYAAPEIVSGIPYQGFASDVWSCGVILFALLT- 226
Query 232 GELPFGLSSLPPDEE-----TVAYVVQLGAHASPESLTVDMEYRGIHKRWQSLIVRLLNP 286
G LPF DEE T+ VQ G P D E I + Q LI ++L
Sbjct 227 GRLPF-------DEEDGNIRTLLLKVQKGEFEMPS----DDE---ISREAQDLIRKILTV 272
Query 287 DREKRISAEEIVK 299
D E+RI +I+K
Sbjct 273 DPERRIKTRDILK 285
> CE05507
Length=785
Score = 40.8 bits (94), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 35/107 (32%), Positives = 56/107 (52%), Gaps = 12/107 (11%)
Query 191 YTEPELLASIVCEESCEFQPIANPKADLWSLGVLLYEILMDGELPFGLSSLPPDEETVAY 250
Y PE+L + + QP N + DLWSLGV+L+ +L G++PF S +E+
Sbjct 566 YAAPEVL------DVGDSQPEYNEQCDLWSLGVVLFTML-SGQVPFHARS---RQESATE 615
Query 251 VVQLGAHASPESLTVDMEYRGIHKRWQSLIVRLLNPDREKRISAEEI 297
++Q A S T D + + ++LI LL D +KR+S +E+
Sbjct 616 IMQRICRAEF-SFTGD-AWTNVSADAKNLITGLLTVDPKKRLSMQEL 660
> Hs16418343
Length=268
Score = 40.4 bits (93), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 50/187 (26%), Positives = 81/187 (43%), Gaps = 30/187 (16%)
Query 119 DVLDWVPTG-----TRA-FLARHVLKTVLHLQWAGWSNNQINSSSFGVQDDGSVLL-FGF 171
DV D V G +RA L R +++ + + G ++ + + +Q L FGF
Sbjct 95 DVFDCVLNGGPLPESRAKALFRQMVEAIRYCHGCGVAHRDLKCENALLQGFNLKLTDFGF 154
Query 172 ESSVPFGSVIPADADV-SAVYTEPELLASIVCEESCEFQPIANPKADLWSLGVLLYEILM 230
+P + S Y PE+L I P + K D+WS+GV+LY +++
Sbjct 155 AKVLPKSHRELSQTFCGSTAYAAPEVLQGI---------PHDSKKGDVWSMGVVLY-VML 204
Query 231 DGELPFGLSSLPPDEETVAYVVQLGAHASPESLTVDMEYRGIHKRWQSLIVRLLNPDREK 290
LPF + +P + + Q G + P L++ + Q L+ RLL PD
Sbjct 205 CASLPFDDTDIP----KMLWQQQKGV-SFPTHLSISADC-------QDLLKRLLEPDMIL 252
Query 291 RISAEEI 297
R S EE+
Sbjct 253 RPSIEEV 259
> CE26793_1
Length=795
Score = 39.3 bits (90), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 58/276 (21%), Positives = 107/276 (38%), Gaps = 56/276 (20%)
Query 68 AAAVAKEKGIAAITSVAKIKGMPDVLHT-QHFTVLSTVEKMEN----IQVT-------LR 115
A + K+ I + + +I+ ++ H ++ E EN I V L
Sbjct 102 AVKLIKKSAIESKADLVRIRREIRIMSALNHPNIIQIYEVFENKDKIILVMEYSSGGELY 161
Query 116 DVLDVLDWVPTGTRAFLARHVLKTVLHLQWAGWSNNQINSSSFGVQDDGSVLLFGFESSV 175
D + +P + R + VL+ ++ + + + + + + F S
Sbjct 162 DYVSRCGSLPEAEARRIFRQITSAVLYCHKHRVAHRDLKLENILLDQNNNAKIADFGLSN 221
Query 176 PFGSVIPADADV------SAVYTEPELLASIVCEESCEFQPIANPKADLWSLGVLLYEIL 229
F AD ++ S +Y PE++ P P+ D WSLG+LLY L
Sbjct 222 YF-----ADKNLLTTFCGSPLYASPEIINGT---------PYKGPEVDCWSLGILLY-TL 266
Query 230 MDGELPFGLSSLPPDEETVAYVVQLGAHASPES-LTVDMEYRGIHKRWQSLIVRLLNPDR 288
+ G +PF D + ++ GA+ PE+ T M LI +L +
Sbjct 267 VYGSMPFDGR----DFNRMVRQIKRGAYFEPETPSTASM-----------LIRNMLRVNP 311
Query 289 EKRISAEEI-------VKDFSDLLNGLPEQQVLDDT 317
E+R + +I +++ ++ LPE Q++D T
Sbjct 312 ERRATIFDIASHWWLNLEENMPVIQELPENQIIDHT 347
> Hs8923922
Length=795
Score = 38.9 bits (89), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 33/137 (24%), Positives = 56/137 (40%), Gaps = 17/137 (12%)
Query 134 RHVLKTVLHLQWAGWSNNQINSSSFGVQDDGSVLL--FGFESSVPFGSVIPADADVSAVY 191
R ++ V + + + + + + D ++ + FGF + G+ + S Y
Sbjct 164 RQIVSAVQYCHQKYIVHRDLKAENLLLDGDMNIKIADFGFSNEFTVGNKLDTFCG-SPPY 222
Query 192 TEPELLASIVCEESCEFQPIANPKADLWSLGVLLYEILMDGELPFGLSSLPPDEETVAYV 251
PEL + P+ D+WSLGV+LY L+ G LPF +L E V
Sbjct 223 AAPELFQG---------KKYDGPEVDVWSLGVILY-TLVSGSLPFDGQNLKELRERVLR- 271
Query 252 VQLGAHASPESLTVDME 268
G + P ++ D E
Sbjct 272 ---GKYRIPFYMSTDCE 285
> At1g74740
Length=567
Score = 37.7 bits (86), Expect = 0.085, Method: Compositional matrix adjust.
Identities = 37/133 (27%), Positives = 67/133 (50%), Gaps = 18/133 (13%)
Query 214 PKADLWSLGVLLYEILMDGELPFGLSSLPPDEETVAYVVQLGAHASPESLTVDME---YR 270
P+ D+WS GV+LY IL+ G PF + E+ VA + G +D + +
Sbjct 238 PEVDVWSAGVILY-ILLCGVPPFWAET----EQGVALAILRG--------VLDFKRDPWS 284
Query 271 GIHKRWQSLIVRLLNPDREKRISAEEIVKDFSDLLNGLPEQQVLDDTTFLDLFLQENPNF 330
I + +SL+ ++L PD KR++A++++ + + +L L E+ T F ++P +
Sbjct 285 QISESAKSLVKQMLEPDSTKRLTAQQVLGN-NAVLVSLMEKMAFKVKTVSVSFFADHP-W 342
Query 331 FREQAEMPNVKRG 343
+ + PNV G
Sbjct 343 IQNAKKAPNVPLG 355
> SPCC18B5.11c
Length=460
Score = 37.7 bits (86), Expect = 0.093, Method: Compositional matrix adjust.
Identities = 32/106 (30%), Positives = 50/106 (47%), Gaps = 14/106 (13%)
Query 191 YTEPELLASIVCEESCEFQPIANPKADLWSLGVLLYEILMDGELPFGLSSLPPDEETVAY 250
Y PE+L S ++ + K D+WSLG +LY +++ +PF SS +
Sbjct 335 YLAPEVLKS----KNVNLDGGYDDKVDIWSLGCVLY-VMLTASIPFASSS----QAKCIE 385
Query 251 VVQLGAHASPESLTVDMEYRGIHKRWQSLIVRLLNPDREKRISAEE 296
++ GA+ L ++ GI LI R+L + EKRIS E
Sbjct 386 LISKGAYPIEPLLENEISEEGI-----DLINRMLEINPEKRISESE 426
> Hs4758016
Length=607
Score = 36.2 bits (82), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 50/203 (24%), Positives = 87/203 (42%), Gaps = 30/203 (14%)
Query 214 PKADLWSLGVLLYEILMDGELPFGLSSLPPDEETVAYVVQLGAHASPESLTVDMEYRGIH 273
P+AD+WSLG ++Y +L G PF + L +ET + Q+ + P SL++
Sbjct 200 PEADVWSLGCVMYTLLC-GSPPFETADL---KETYRCIKQV-HYTLPASLSLPA------ 248
Query 274 KRWQSLIVRLLNPDREKRISAEEIVKDFSDLLNGLPEQQVLDDTTFLDLFLQENPNFFRE 333
+ L+ +L R S ++I++ P++ + + NP
Sbjct 249 ---RQLLAAILRASPRDRPSIDQILRHDFFTKGYTPDRLPISSCVTVPDLTPPNP----A 301
Query 334 QAEMPNVKRGLMGEQRPSSPDERQEREDVAGAASGAA-------PATPASPTGAEPRPTR 386
++ V + L G ++ S + QER++V+G SG A P +P + P P
Sbjct 302 RSLFAKVTKSLFGRKK-KSKNHAQERDEVSGLVSGLMRTSVGHQDARPEAPAASGPAPVS 360
Query 387 PQELPLQAPKTDEPVRSTVRTEG 409
E AP+ P R T+ + G
Sbjct 361 LVE---TAPEDSSP-RGTLASSG 379
> Hs13129008
Length=501
Score = 32.7 bits (73), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 22/87 (25%), Positives = 40/87 (45%), Gaps = 12/87 (13%)
Query 217 DLWSLGVLLYEILMDGELPFGLSSLPPDEETVAYVVQLGAHASPESLTVDMEYRG----- 271
D W++GV++Y IL+ G PF + + L D E+
Sbjct 202 DCWAIGVIMY-ILLSGNPPFYEEV------EEDDYENHDKNLFRKILAGDYEFDSPYWDD 254
Query 272 IHKRWQSLIVRLLNPDREKRISAEEIV 298
I + + L+ RL+ ++++RI+AEE +
Sbjct 255 ISQAAKDLVTRLMEVEQDQRITAEEAI 281
Lambda K H
0.311 0.129 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 21604309048
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40