bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0578_orf8
Length=735
Score E
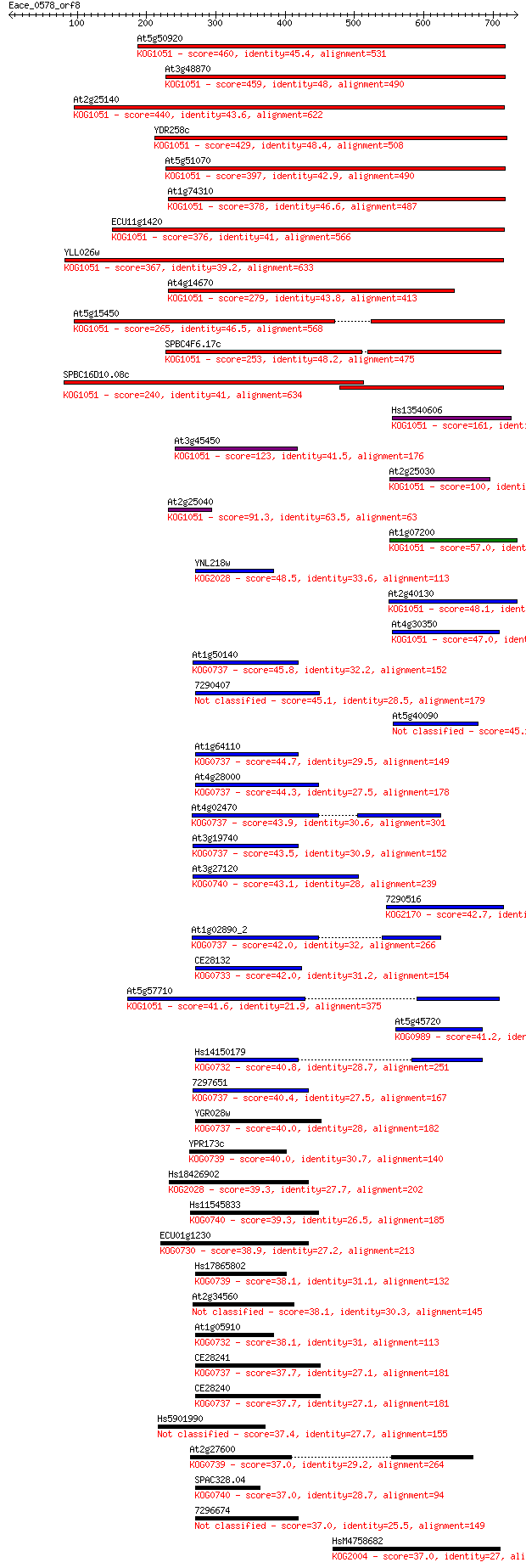
Sequences producing significant alignments: (Bits) Value
At5g50920 460 4e-129
At3g48870 459 1e-128
At2g25140 440 5e-123
YDR258c 429 1e-119
At5g51070 397 4e-110
At1g74310 378 2e-104
ECU11g1420 376 8e-104
YLL026w 367 5e-101
At4g14670 279 2e-74
At5g15450 265 2e-70
SPBC4F6.17c 253 8e-67
SPBC16D10.08c 240 9e-63
Hs13540606 161 4e-39
At3g45450 123 1e-27
At2g25030 100 1e-20
At2g25040 91.3 7e-18
At1g07200 57.0 1e-07
YNL218w 48.5 6e-05
At2g40130 48.1 7e-05
At4g30350 47.0 2e-04
At1g50140 45.8 3e-04
7290407 45.1 6e-04
At5g40090 45.1 7e-04
At1g64110 44.7 8e-04
At4g28000 44.3 0.001
At4g02470 43.9 0.001
At3g19740 43.5 0.002
At3g27120 43.1 0.003
7290516 42.7 0.003
At1g02890_2 42.0 0.005
CE28132 42.0 0.005
At5g57710 41.6 0.006
At5g45720 41.2 0.009
Hs14150179 40.8 0.011
7297651 40.4 0.013
YGR028w 40.0 0.019
YPR173c 40.0 0.020
Hs18426902 39.3 0.030
Hs11545833 39.3 0.034
ECU01g1230 38.9 0.037
Hs17865802 38.1 0.063
At2g34560 38.1 0.074
At1g05910 38.1 0.081
CE28241 37.7 0.091
CE28240 37.7 0.096
Hs5901990 37.4 0.12
At2g27600 37.0 0.15
SPAC328.04 37.0 0.17
7296674 37.0 0.17
HsM4758682 37.0 0.17
> At5g50920
Length=929
Score = 460 bits (1184), Expect = 4e-129, Method: Compositional matrix adjust.
Identities = 241/555 (43%), Positives = 359/555 (64%), Gaps = 25/555 (4%)
Query 187 KILVNSGCDVELFKEE-LSLAYKNTPMPNTSGTTEQLTKDFDITQYGEDLTEKAAKGKLQ 245
++L N G D + + + + +N + G K + +YG +LT+ A +GKL
Sbjct 215 RVLENLGADPSNIRTQVIRMVGENNEVTANVGGGSSSNKMPTLEEYGTNLTKLAEEGKLD 274
Query 246 PVIGRDEEIDRIAQILSRMMTKAPLLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRII 305
PV+GR +I+R+ QIL R P LIGEPGVGKTA+ EGLAQRI G VP+ + G+++I
Sbjct 275 PVVGRQPQIERVVQILGRRTKNNPCLIGEPGVGKTAIAEGLAQRIASGDVPETIEGKKVI 334
Query 306 SLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQI 365
+LD+ L AGT RGEFE+R+K+++ + R+ ++ILF+DE+HTL+GAG G+ DA I
Sbjct 335 TLDMGLLVAGTKYRGEFEERLKKLMEEI-RQSDEIILFIDEVHTLIGAGAAEGAIDAANI 393
Query 366 LKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYE 425
LK LARGE+ +GATTL+EY+ HIEKD A RRFQ + V P+ + + ILK ++ YE
Sbjct 394 LKPALARGELQCIGATTLDEYRKHIEKDPALERRFQPVKVPEPTVDETIQILKGLRERYE 453
Query 426 KFHSVDLSDEVLEGIVALSDQYVKQRSFPDKALDLLDEACSWKKVSH----------NKQ 475
H + +DE L LS QY+ R PDKA+DL+DEA S ++ H K+
Sbjct 454 IHHKLRYTDESLVAAAQLSYQYISDRFLPDKAIDLIDEAGSRVRLRHAQVPEEARELEKE 513
Query 476 IVVLKRKIEEAGKKKDETAAEELQKWKDEL-AALEELTANGRRLA------------LSL 522
+ + ++ EA + +D A L+ + EL A + + A G+ ++ ++
Sbjct 514 LRQITKEKNEAVRGQDFEKAGTLRDREIELRAEVSAIQAKGKEMSKAESETGEEGPMVTE 573
Query 523 HDVAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSP 582
D+ HI+ WTGIP+ K++ DE R+L++ + L R+IGQ +AVKA++ A+ R GL
Sbjct 574 SDIQHIVSSWTGIPVEKVSTDESDRLLKMEETLHKRIIGQDEAVKAISRAIRRARVGLKN 633
Query 583 KNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNLIRLDMVEYQEAHSISRLIGPPPGY 642
N+P+ +F+F G +GVGK+ELAKA+A F SE+ +IRLDM E+ E H++S+LIG PPGY
Sbjct 634 PNRPIASFIFSGPTGVGKSELAKALAAYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGY 693
Query 643 MGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCL 702
+G EGGQLTEAVR++P++VVLFDE+E AH ++++++L +L++G L+DSKG VDF N L
Sbjct 694 VGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTL 753
Query 703 IILTSNIGRQHILEG 717
+I+TSN+G I +G
Sbjct 754 LIMTSNVGSSVIEKG 768
> At3g48870
Length=952
Score = 459 bits (1180), Expect = 1e-128, Method: Compositional matrix adjust.
Identities = 235/513 (45%), Positives = 345/513 (67%), Gaps = 24/513 (4%)
Query 228 ITQYGEDLTEKAAKGKLQPVIGRDEEIDRIAQILSRMMTKAPLLIGEPGVGKTAVIEGLA 287
+ +YG +LT+ A +GKL PV+GR +I+R+ QIL+R P LIGEPGVGKTA+ EGLA
Sbjct 278 LEEYGTNLTKLAEEGKLDPVVGRQPQIERMVQILARRTKNNPCLIGEPGVGKTAIAEGLA 337
Query 288 QRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEI 347
QRI G VP+ + G+ +I+LD+ L AGT RGEFE+R+K+++ + R+ ++ILF+DE+
Sbjct 338 QRIASGDVPETIEGKTVITLDMGLLVAGTKYRGEFEERLKKLMEEI-RQSDEIILFIDEV 396
Query 348 HTLVGAGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEA 407
HTL+GAG G+ DA ILK LARGE+ +GATT++EY+ HIEKD A RRFQ + V
Sbjct 397 HTLIGAGAAEGAIDAANILKPALARGELQCIGATTIDEYRKHIEKDPALERRFQPVKVPE 456
Query 408 PSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQRSFPDKALDLLDEACSW 467
P+ E A+ IL+ ++ YE H + +DE L LS QY+ R PDKA+DL+DEA S
Sbjct 457 PTVEEAIQILQGLRERYEIHHKLRYTDEALVAAAQLSHQYISDRFLPDKAIDLIDEAGSR 516
Query 468 KKVSH----------NKQIVVLKRKIEEAGKKKDETAAEELQKWKDELAA-LEELTANGR 516
++ H KQ+ + ++ EA + +D A + + EL A + + + G+
Sbjct 517 VRLRHAQLPEEARELEKQLRQITKEKNEAVRSQDFEMAGSHRDREIELKAEIANVLSRGK 576
Query 517 RLA------------LSLHDVAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQ 564
+A ++ D+ HI+ WTGIP+ K++ DE SR+L++ L +RVIGQ +
Sbjct 577 EVAKAENEAEEGGPTVTESDIQHIVATWTGIPVEKVSSDESSRLLQMEQTLHTRVIGQDE 636
Query 565 AVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNLIRLDMV 624
AVKA++ A+ R GL N+P+ +F+F G +GVGK+ELAKA+A F SE+ +IRLDM
Sbjct 637 AVKAISRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKALAAYYFGSEEAMIRLDMS 696
Query 625 EYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPMLD 684
E+ E H++S+LIG PPGY+G EGGQLTEAVR++P+++VLFDE+E AH ++++++L +L+
Sbjct 697 EFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTLVLFDEIEKAHPDVFNMMLQILE 756
Query 685 EGHLSDSKGNRVDFTNCLIILTSNIGRQHILEG 717
+G L+DSKG VDF N L+I+TSN+G I +G
Sbjct 757 DGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKG 789
> At2g25140
Length=874
Score = 440 bits (1132), Expect = 5e-123, Method: Compositional matrix adjust.
Identities = 271/707 (38%), Positives = 396/707 (56%), Gaps = 94/707 (13%)
Query 95 HLLMVLLEGSRLDTPREVITKCHGDFSHVRGAVKAVLVRLPRTPVTVPLGGAKVTPAVTE 154
HL+ LLE + R++ TK D S V ++A + + + P G ++ +++
Sbjct 25 HLMKALLE-QKDGMARKIFTKAGIDNSSV---LQATDLFISKQPTVSDASGQRLGSSLSV 80
Query 155 VFLRARR-IAEAEGSLTGVQHVFLALLYCPTFY--KILVNSGCDVELFKEELSLAYKNTP 211
+ A+R + S V+H LA Y T + + + D+++ K+ A K+
Sbjct 81 ILENAKRHKKDMLDSYVSVEHFLLAY-YSDTRFGQEFFRDMKLDIQVLKD----AIKDVR 135
Query 212 MPNTSGTTEQLTKDFDITQYGEDLTEKAAKGKLQPVIGRDEEIDRIAQILSRMMTKAPLL 271
+K + +YG DLTE A +GKL PVIGRD+EI R QIL R P++
Sbjct 136 GDQRVTDRNPESKYQALEKYGNDLTEMARRGKLDPVIGRDDEIRRCIQILCRRTKNNPVI 195
Query 272 IGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIIT 331
IGEPGVGKTA+ EGLAQRIV G VP+ L R++ISLD+ +L AG RG+FE+R+K ++
Sbjct 196 IGEPGVGKTAIAEGLAQRIVRGDVPEPLMNRKLISLDMGSLLAGAKFRGDFEERLKAVMK 255
Query 332 FLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIE 391
+ ILF+DEIHT+VGAG G+ DA+ +LK L RGE+ +GATTL EY+ +IE
Sbjct 256 EVSASNGQTILFIDEIHTVVGAGAMDGAMDASNLLKPMLGRGELRCIGATTLTEYRKYIE 315
Query 392 KDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFH----------------------- 428
KD A RRFQ ++ PS E +SIL+ ++ YE H
Sbjct 316 KDPALERRFQQVLCVQPSVEDTISILRGLRERYELHHGVTISDSALVSAAVLADRYITER 375
Query 429 -----SVDLSDEV--------------LEGI----VALSDQYVKQRSFPDKA-------- 457
++DL DE L+GI + L + + ++ DKA
Sbjct 376 FLPDKAIDLVDEAGAKLKMEITSKPTELDGIDRAVIKLEMEKLSLKNDTDKASKERLQKI 435
Query 458 ---LDLLDEA-----CSWKK--------VSHNKQIVVLKRKIEEAGKKKDETAAEELQ-- 499
L L + W+K S ++I + +IE A ++ D A EL+
Sbjct 436 ENDLSTLKQKQKELNVQWEKEKSLMTKIRSFKEEIDRVNLEIESAEREYDLNRAAELKYG 495
Query 500 ---KWKDELAALEELTANGRRLALSLH-------DVAHILHLWTGIPLGKMTEDEISRVL 549
+ +L E+ N R+ SL D+A I+ WTGIPL + + E +++
Sbjct 496 TLLSLQRQLEEAEKNLTNFRQFGQSLLREVVTDLDIAEIVSKWTGIPLSNLQQSEREKLV 555
Query 550 RLADILSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAE 609
L ++L RVIGQ AVK+VADA+ RAGLS N+P+ +FMF+G +GVGKTELAKA+A
Sbjct 556 MLEEVLHHRVIGQDMAVKSVADAIRRSRAGLSDPNRPIASFMFMGPTGVGKTELAKALAG 615
Query 610 EMFDSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVE 669
+F++E ++R+DM EY E HS+SRL+G PPGY+G +EGGQLTE VR++P+SVVLFDE+E
Sbjct 616 YLFNTENAIVRVDMSEYMEKHSVSRLVGAPPGYVGYEEGGQLTEVVRRRPYSVVLFDEIE 675
Query 670 NAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGRQHILE 716
AH +++++LL +LD+G ++DS+G V F NC++I+TSNIG HILE
Sbjct 676 KAHPDVFNILLQLLDDGRITDSQGRTVSFKNCVVIMTSNIGSHHILE 722
> YDR258c
Length=811
Score = 429 bits (1104), Expect = 1e-119, Method: Compositional matrix adjust.
Identities = 246/594 (41%), Positives = 354/594 (59%), Gaps = 88/594 (14%)
Query 212 MPNTSGTTEQLTKDFD-------ITQYGEDLTEKAAKGKLQPVIGRDEEIDRIAQILSRM 264
+PN T Q+ D + + Q+G +LT+ A GKL PVIGRDEEI R QILSR
Sbjct 75 LPNFKRTYVQMRMDPNQQPEKPALEQFGTNLTKLARDGKLDPVIGRDEEIARAIQILSRR 134
Query 265 MTKAPLLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEK 324
P LIG GVGKTA+I+GLAQRIV G VP L+ + +++LDL +L AG RGEFE+
Sbjct 135 TKNNPCLIGRAGVGKTALIDGLAQRIVAGEVPDSLKDKDLVALDLGSLIAGAKYRGEFEE 194
Query 325 RMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLARGEIVLVGATTLE 384
R+K+++ + + VI+F+DE+H L+G GKT GS DA+ ILK LARG + + ATTL+
Sbjct 195 RLKKVLEEIDKANGKVIVFIDEVHMLLGLGKTDGSMDASNILKPKLARG-LRCISATTLD 253
Query 385 EYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALS 444
E+K+ IEKD A RRFQ I++ PS +SIL+ +K YE H V ++D L LS
Sbjct 254 EFKI-IEKDPALSRRFQPILLNEPSVSDTISILRGLKERYEVHHGVRITDTALVSAAVLS 312
Query 445 DQYVKQRSFPDKALDLLDEACSWKKVSH----------NKQIVVLKRKIEEAGKKKDETA 494
++Y+ R PDKA+DL+DEAC+ ++ H ++ I+ ++ ++E K+ D +
Sbjct 313 NRYITDRFLPDKAIDLVDEACAVLRLQHESKPDEIQKLDRAIMKIQIELESLKKETDPVS 372
Query 495 AE--------------ELQK----WKDELAALEEL---TAN------------------- 514
E EL + W E A +E + AN
Sbjct 373 VERREALEKDLEMKNDELNRLTKIWDAERAEIESIKNAKANLEQARIELEKCQREGDYTK 432
Query 515 ------------GRRLALS-----------LHD------VAHILHLWTGIPLGKMTEDEI 545
+++ALS LHD ++ ++ TGIP + + +
Sbjct 433 ASELRYSRIPDLEKKVALSEKSKDGDKVNLLHDSVTSDDISKVVAKMTGIPTETVMKGDK 492
Query 546 SRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAK 605
R+L + + L RV+GQ +A+ A++DA+ +QRAGL+ + +P+ +FMFLG +G GKTEL K
Sbjct 493 DRLLYMENSLKERVVGQDEAIAAISDAVRLQRAGLTSEKRPIASFMFLGPTGTGKTELTK 552
Query 606 AVAEEMFDSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLF 665
A+AE +FD E N+IR DM E+QE H++SRLIG PPGY+ ++ GGQLTEAVR+KP++VVLF
Sbjct 553 ALAEFLFDDESNVIRFDMSEFQEKHTVSRLIGAPPGYVLSESGGQLTEAVRRKPYAVVLF 612
Query 666 DEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGRQHILEGYK 719
DE E AH ++ +LL +LDEG L+DS G+ VDF N +I++TSNIG+ +L K
Sbjct 613 DEFEKAHPDVSKLLLQVLDEGKLTDSLGHHVDFRNTIIVMTSNIGQDILLNDTK 666
> At5g51070
Length=945
Score = 397 bits (1021), Expect = 4e-110, Method: Compositional matrix adjust.
Identities = 210/523 (40%), Positives = 322/523 (61%), Gaps = 39/523 (7%)
Query 228 ITQYGEDLTEKAAKGKLQPVIGRDEEIDRIAQILSRMMTKAPLLIGEPGVGKTAVIEGLA 287
+ Q+ DLT +A++G + PVIGR++E+ R+ QIL R P+L+GE GVGKTA+ EGLA
Sbjct 271 LEQFCVDLTARASEGLIDPVIGREKEVQRVIQILCRRTKNNPILLGEAGVGKTAIAEGLA 330
Query 288 QRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEI 347
I E + P L +RI+SLD+ L AG RGE E R+ +I+ +++ K VILF+DE+
Sbjct 331 ISIAEASAPGFLLTKRIMSLDIGLLMAGAKERGELEARVTALISEVKKSGK-VILFIDEV 389
Query 348 HTLVGAGKTS----GSS-DATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQN 402
HTL+G+G GS D +LK L RGE+ + +TTL+E++ EKD A RRFQ
Sbjct 390 HTLIGSGTVGRGNKGSGLDIANLLKPSLGRGELQCIASTTLDEFRSQFEKDKALARRFQP 449
Query 403 IVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQRSFPDKALDLLD 462
+++ PS+E A+ IL ++ YE H+ + E ++ V LS +Y+ R PDKA+DL+D
Sbjct 450 VLINEPSEEDAVKILLGLREKYEAHHNCKYTMEAIDAAVYLSSRYIADRFLPDKAIDLID 509
Query 463 EACS-------------------------W---KKVSHNKQIVVLKRKIEEAGKKKDETA 494
EA S W K V ++V+ R+ ++ G + +
Sbjct 510 EAGSRARIEAFRKKKEDAICILSKPPNDYWQEIKTVQAMHEVVLSSRQKQDDGDAISDES 569
Query 495 AEELQKWKDELAALEELTANGRRLALSLHDVAHILHLWTGIPLGKMTEDEISRVLRLADI 554
E +++ ++L + + + D+A + +W+GIP+ ++T DE ++ L D
Sbjct 570 GELVEE-----SSLPPAAGDDEPILVGPDDIAAVASVWSGIPVQQITADERMLLMSLEDQ 624
Query 555 LSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDS 614
L RV+GQ +AV A++ A+ R GL ++P+ +F G +GVGKTEL KA+A F S
Sbjct 625 LRGRVVGQDEAVAAISRAVKRSRVGLKDPDRPIAAMLFCGPTGVGKTELTKALAANYFGS 684
Query 615 EKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKN 674
E++++RLDM EY E H++S+LIG PPGY+G +EGG LTEA+R++P +VVLFDE+E AH +
Sbjct 685 EESMLRLDMSEYMERHTVSKLIGSPPGYVGFEEGGMLTEAIRRRPFTVVLFDEIEKAHPD 744
Query 675 LWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGRQHILEG 717
++++LL + ++GHL+DS+G RV F N LII+TSN+G I +G
Sbjct 745 IFNILLQLFEDGHLTDSQGRRVSFKNALIIMTSNVGSLAIAKG 787
> At1g74310
Length=911
Score = 378 bits (971), Expect = 2e-104, Method: Compositional matrix adjust.
Identities = 227/566 (40%), Positives = 317/566 (56%), Gaps = 82/566 (14%)
Query 231 YGEDLTEKAAKGKLQPVIGRDEEIDRIAQILSRMMTKAPLLIGEPGVGKTAVIEGLAQRI 290
YG DL E+A GKL PVIGRDEEI R+ +ILSR P+LIGEPGVGKTAV+EGLAQRI
Sbjct 167 YGRDLVEQA--GKLDPVIGRDEEIRRVVRILSRRTKNNPVLIGEPGVGKTAVVEGLAQRI 224
Query 291 VEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTL 350
V+G VP L R+ISLD+ L AG RGEFE+R+K ++ ++ + VILF+DEIH +
Sbjct 225 VKGDVPNSLTDVRLISLDMGALVAGAKYRGEFEERLKSVLKEVEDAEGKVILFIDEIHLV 284
Query 351 VGAGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSK 410
+GAGKT GS DA + K LARG++ +GATTLEEY+ ++EKDAAF RRFQ + V PS
Sbjct 285 LGAGKTEGSMDAANLFKPMLARGQLRCIGATTLEEYRKYVEKDAAFERRFQQVYVAEPSV 344
Query 411 ERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQRSFPDKALDLLDEACSWKKV 470
+SIL+ +K YE H V + D L LS +Y+ R PDKA+DL+DEAC+ +V
Sbjct 345 PDTISILRGLKEKYEGHHGVRIQDRALINAAQLSARYITGRHLPDKAIDLVDEACANVRV 404
Query 471 SHNKQ-----------------IVVLKRKIEEAGKKKDETAAEELQKWKDELAAL----- 508
+ Q + L+R+ ++A K + +EL +D+L L
Sbjct 405 QLDSQPEEIDNLERKRMQLEIELHALEREKDKASKARLIEVRKELDDLRDKLQPLTMKYR 464
Query 509 ---------EELTANGRRLALSLHDVAHILHLWTGIPL--GKMTEDEISRVLRL------ 551
L L SL + L L G + E E S + +L
Sbjct 465 KEKERIDEIRRLKQKREELMFSLQEAERRYDLARAADLRYGAIQEVE-SAIAQLEGTSSE 523
Query 552 ADILSSRVIGQQQAVKAVA--DALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAE 609
+++ + +G + + V+ + + R G + K + +G L VG+ + AV+E
Sbjct 524 ENVMLTENVGPEHIAEVVSRWTGIPVTRLGQNEKERLIGLADRLHKRVVGQNQAVNAVSE 583
Query 610 --------------------------------------EMFDSEKNLIRLDMVEYQEAHS 631
++FD E L+R+DM EY E HS
Sbjct 584 AILRSRAGLGRPQQPTGSFLFLGPTGVGKTELAKALAEQLFDDENLLVRIDMSEYMEQHS 643
Query 632 ISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPMLDEGHLSDS 691
+SRLIG PPGY+G++EGGQLTEAVR++P+ V+LFDEVE AH +++ LL +LD+G L+D
Sbjct 644 VSRLIGAPPGYVGHEEGGQLTEAVRRRPYCVILFDEVEKAHVAVFNTLLQVLDDGRLTDG 703
Query 692 KGNRVDFTNCLIILTSNIGRQHILEG 717
+G VDF N +II+TSN+G +H+L G
Sbjct 704 QGRTVDFRNSVIIMTSNLGAEHLLAG 729
> ECU11g1420
Length=851
Score = 376 bits (966), Expect = 8e-104, Method: Compositional matrix adjust.
Identities = 232/645 (35%), Positives = 357/645 (55%), Gaps = 91/645 (14%)
Query 150 PAVTEVFLRARRIAEA--EGSLTGVQHVFLALLYCPTFYKILVNSGCDVELFKEELSLAY 207
P V E + AE+ EG V + L+LL + +++ NS E ++++
Sbjct 78 PRVGETLAKVLMAAESKGEGEYVSVNALVLSLLEVESIRELIANS----EDIRKKVEEQT 133
Query 208 KNTPMP--NTSGTTEQLTKDFDITQYGEDLTEKAAKGKLQPVIGRDEEIDRIAQILSRMM 265
KN N T+ +++ + D+ E+A + PVIGRDEEI R+ +ILS+
Sbjct 134 KNKRFDSRNADDGTDVMSR------FAVDMVEQARQNVFDPVIGRDEEIRRVLEILSKKT 187
Query 266 TKAPLLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKR 325
+L+G+PGVGKTA++ G+AQ I G P LRG RI ++D+ ++ AGTS RG+FE+R
Sbjct 188 KSNAILVGKPGVGKTAIVNGIAQLIANGEAPT-LRGARIYNVDVGSMVAGTSHRGDFEER 246
Query 326 MKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLARGEIVLVGATTLEE 385
+K ++ + VILF+DEIH ++GAGKT G+ DA +LK LA G I +GATT +E
Sbjct 247 LKTLVKEAET-TPGVILFIDEIHIVLGAGKTDGAMDAANMLKPGLAAGTIKCIGATTHDE 305
Query 386 YKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSD 445
Y+ +IE D AF RRF +VV PS E ++++L+ +K E +H V ++D + V S
Sbjct 306 YRKYIENDPAFERRFVQVVVNEPSIEDSITMLRGLKGRLEAYHGVKIADSAIVYAVNSSK 365
Query 446 QYVKQRSFPDKALDLLDEACSWKKV---SHNKQIV----------VLKRKIE-EAGKKKD 491
+Y+ R PD A+DLLD AC+ + S ++I+ + K +E + +KKD
Sbjct 366 KYISNRRLPDVAIDLLDTACASVMIALESEPQEILNQKSKAWGLDIAKTSLEFDLSQKKD 425
Query 492 ET-------AAEELQKWKDELAALEELTANGR------------------RLA------- 519
E EEL+K K L +EE + RLA
Sbjct 426 EQTLKKLKEVVEELEKVKASLQPMEEAYLRDKKHIIEAKKLKKKLEDTKLRLAQAERDRN 485
Query 520 ---------------------LSLHDVAHIL--HL------WTGIPLGKMTEDEISRVLR 550
L L V IL H+ WTGI + ++T E R++
Sbjct 486 TYVAYDLKTNVIPVIESELKRLELESVVVILPSHVAEIISRWTGIDVKRLTIKENERLME 545
Query 551 LADILSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEE 610
++ + R+ GQ AV A+ D++ R GL ++P+G+F+ LG +GVGKTELAKAVA E
Sbjct 546 MSSRIKKRIFGQDHAVDAIVDSILQSRVGLDDDDRPVGSFLLLGPTGVGKTELAKAVAME 605
Query 611 MFDSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVEN 670
+FD+EK+++ +DM EY I++LIG GY+G ++GG LTE ++ +P++V+L DEV+
Sbjct 606 LFDNEKDMLVIDMSEYGNEMGITKLIGANAGYVGYNQGGTLTEPIKGRPYNVILLDEVDL 665
Query 671 AHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGRQHIL 715
AH+ + + L +LDEG ++D KG VDF NC++I+TSN+G++ I+
Sbjct 666 AHQTVLNTLYQLLDEGRVTDGKGAVVDFRNCVVIMTSNLGQEIIM 710
> YLL026w
Length=908
Score = 367 bits (942), Expect = 5e-101, Method: Compositional matrix adjust.
Identities = 248/730 (33%), Positives = 391/730 (53%), Gaps = 113/730 (15%)
Query 82 LAWHLHHDRLCLAHLLMVLLEGSRLDTPRE--------VITKCHGDFSHVRGAVKAVLVR 133
LA H +L H+L +E TP + +I K D+ + V LVR
Sbjct 21 LASDHQHPQLQPIHILAAFIE-----TPEDGSVPYLQNLIEKGRYDYDLFKKVVNRNLVR 75
Query 134 LPRTPVTVPLGGAKVTP--AVTEVFLRARRIAEAE-GSLTGVQHVFLALLYCPTFYKILV 190
+P+ P A++TP A+ +V A +I + + S H+ AL + +I
Sbjct 76 IPQQQ-PAP---AEITPSYALGKVLQDAAKIQKQQKDSFIAQDHILFALFNDSSIQQIFK 131
Query 191 NSGCDVELFKEELSLAYKNTPMPNTSGTTEQLTKDFDITQYGEDLTEKAAKGKLQPVIGR 250
+ D+E K++ +L + ++ G ++ +++Y D+TE+A +GKL PVIGR
Sbjct 132 EAQVDIEAIKQQ-ALELRGNTRIDSRGADTNTPLEY-LSKYAIDMTEQARQGKLDPVIGR 189
Query 251 DEEIDRIAQILSRMMTKAPLLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLL 310
+EEI ++L+R + P LIGEPG+GKTA+IEG+AQRI++ VP L+G ++ SLDL
Sbjct 190 EEEIRSTIRVLARRIKSNPCLIGEPGIGKTAIIEGVAQRIIDDDVPTILQGAKLFSLDLA 249
Query 311 NLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPL 370
L+AG +G+FE+R K ++ ++ K ++LF+DEIH L+G GK DA ILK L
Sbjct 250 ALTAGAKYKGDFEERFKGVLKEIEESKTLIVLFIDEIHMLMGNGK----DDAANILKPAL 305
Query 371 ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFHSV 430
+RG++ ++GATT EY+ +EKD AF RRFQ I V PS + ++IL+ ++ YE H V
Sbjct 306 SRGQLKVIGATTNNEYRSIVEKDGAFERRFQKIEVAEPSVRQTVAILRGLQPKYEIHHGV 365
Query 431 DLSDEVLEGIVALSDQYVKQRSFPDKALDLLDEACSWKKV----------SHNKQIVVLK 480
+ D L L+ +Y+ R PD ALDL+D +C+ V S +Q+ +++
Sbjct 366 RILDSALVTAAQLAKRYLPYRRLPDSALDLVDISCAGVAVARDSKPEELDSKERQLQLIQ 425
Query 481 RKI------EEA----------GKKKDETAAEEL----QKWKDELAALEELTANGRRL-- 518
+I E+A ++K+ + EEL Q++ +E EELT ++L
Sbjct 426 VEIKALERDEDADSTTKDRLKLARQKEASLQEELEPLRQRYNEEKHGHEELTQAKKKLDE 485
Query 519 -------ALSLHDVAHILHL-WTGIPLGKMT----EDEISRVLRLA------------DI 554
A +D A L + IP K ED+++ R A D
Sbjct 486 LENKALDAERRYDTATAADLRYFAIPDIKKQIEKLEDQVAEEERRAGANSMIQNVVDSDT 545
Query 555 LS---SRVIG---------------------------QQQAVKAVADALAIQRAGLSPKN 584
+S +R+ G Q A+KAV++A+ + R+GL+
Sbjct 546 ISETAARLTGIPVKKLSESENEKLIHMERDLSSEVVGQMDAIKAVSNAVRLSRSGLANPR 605
Query 585 KPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNLIRLDMVEYQEAHSISRLIGPPPGYMG 644
+P +F+FLG SG GKTELAK VA +F+ E +IR+D E E +++S+L+G GY+G
Sbjct 606 QP-ASFLFLGLSGSGKTELAKKVAGFLFNDEDMMIRVDCSELSEKYAVSKLLGTTAGYVG 664
Query 645 NDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCLII 704
DEGG LT ++ KP+SV+LFDEVE AH ++ +V+L MLD+G ++ +G +D +NC++I
Sbjct 665 YDEGGFLTNQLQYKPYSVLLFDEVEKAHPDVLTVMLQMLDDGRITSGQGKTIDCSNCIVI 724
Query 705 LTSNIGRQHI 714
+TSN+G + I
Sbjct 725 MTSNLGAEFI 734
> At4g14670
Length=623
Score = 279 bits (713), Expect = 2e-74, Method: Compositional matrix adjust.
Identities = 181/493 (36%), Positives = 266/493 (53%), Gaps = 84/493 (17%)
Query 231 YGEDLTEKAAKGKLQPVIGRDEEIDRIAQILSRMMTKAPLLIGEPGVGKTAVIEGLAQRI 290
YG DL E+A GKL PVIGR EI R+ ++LSR P+LIGEPGVGKTAV+EGLAQRI
Sbjct 132 YGTDLVEQA--GKLDPVIGRHREIRRVIEVLSRRTKNNPVLIGEPGVGKTAVVEGLAQRI 189
Query 291 VEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTL 350
++G VP L G ++ISL+ + AGT++RG+FE+R+K ++ ++ + V+LF+DEIH
Sbjct 190 LKGDVPINLTGVKLISLEFGAMVAGTTLRGQFEERLKSVLKAVEEAQGKVVLFIDEIHMA 249
Query 351 VGAGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSK 410
+GA K SGS+DA ++LK LARG++ +GATTLEEY+ H+EKDAAF RRFQ + V PS
Sbjct 250 LGACKASGSTDAAKLLKPMLARGQLRFIGATTLEEYRTHVEKDAAFERRFQQVFVAEPSV 309
Query 411 ERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQRSFPDKALDLLDEACSWKKV 470
+SIL+ +K YE H V + D L LS++Y+ R PDKA+DL+DE+C+ K
Sbjct 310 PDTISILRGLKEKYEGHHGVRIQDRALVLSAQLSERYITGRRLPDKAIDLVDESCAHVKA 369
Query 471 ----------SHNKQIVVLKRKIEEAGKKKDETAAE--------ELQKWKDELAAL---- 508
S ++++ L+ +I K+KD+ A+E EL +D+L L
Sbjct 370 QLDIQPEEIDSLERKVMQLEIEIHALEKEKDDKASEARLSEVRKELDDLRDKLEPLTIKY 429
Query 509 ----------EELTANGRRLALSL------HDVAHILHLWTGIPLGKMTEDEISRVLRLA 552
L N L ++L HDV L G + E I+++ + A
Sbjct 430 KKEKKIINETRRLKQNRDDLMIALQEAERQHDVPKAAVLKYGAI--QEVESAIAKLEKSA 487
Query 553 --DILSSRVIGQQQAVKAVA--DALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAV- 607
+++ + +G + + V+ + + R + K + + L VG+ E KAV
Sbjct 488 KDNVMLTETVGPENIAEVVSRWTGIPVTRLDQNEKKRLISLADKLHERVVGQDEAVKAVA 547
Query 608 -------------------------------------AEEMFDSEKNLIRLDMVEYQEAH 630
AE++FDSE L+RLDM EY +
Sbjct 548 AAILRSRVGLGRPQQPSGSFLFLGPTGVGKTELAKALAEQLFDSENLLVRLDMSEYNDKF 607
Query 631 SISRLIGPPPGYM 643
S+++LIG PPGY+
Sbjct 608 SVNKLIGAPPGYV 620
> At5g15450
Length=968
Score = 265 bits (678), Expect = 2e-70, Method: Compositional matrix adjust.
Identities = 159/383 (41%), Positives = 228/383 (59%), Gaps = 19/383 (4%)
Query 95 HLLMVLLEGSRLDTPREVITKCHGDFSHVRGAVKAVLVRLPRTPVTVPLGGAKVTPAVTE 154
HL+ LLE + R + +K D + V A + + R P+ V G+ + +
Sbjct 109 HLMKALLE-QKNGLARRIFSKIGVDNTKVLEATEKFIQRQPK--VYGDAAGSMLGRDLEA 165
Query 155 VFLRARRIA-EAEGSLTGVQHVFLALLYCPTFYKILVNSGCDVELFKE-ELSLAYKNTPM 212
+F RAR+ + + S V+H+ LA F K +LFK+ ++S + +
Sbjct 166 LFQRARQFKKDLKDSYVSVEHLVLAFADDKRFGK---------QLFKDFQISERSLKSAI 216
Query 213 PNTSGTTEQLTKDFD-----ITQYGEDLTEKAAKGKLQPVIGRDEEIDRIAQILSRMMTK 267
+ G + +D + + +YG+DLT A +GKL PVIGRD+EI R QILSR
Sbjct 217 ESIRGKQSVIDQDPEGKYEALEKYGKDLTAMAREGKLDPVIGRDDEIRRCIQILSRRTKN 276
Query 268 APLLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMK 327
P+LIGEPGVGKTA+ EGLAQRIV+G VP+ L R++ISLD+ L AG RGEFE R+K
Sbjct 277 NPVLIGEPGVGKTAISEGLAQRIVQGDVPQALMNRKLISLDMGALIAGAKYRGEFEDRLK 336
Query 328 EIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLARGEIVLVGATTLEEYK 387
++ + + +ILF+DEIHT+VGAG T+G+ DA +LK L RGE+ +GATTL+EY+
Sbjct 337 AVLKEVTDSEGQIILFIDEIHTVVGAGATNGAMDAGNLLKPMLGRGELRCIGATTLDEYR 396
Query 388 LHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQY 447
+IEKD A RRFQ + V+ P+ E +SIL+ ++ YE H V +SD L LSD+Y
Sbjct 397 KYIEKDPALERRFQQVYVDQPTVEDTISILRGLRERYELHHGVRISDSALVEAAILSDRY 456
Query 448 VKQRSFPDKALDLLDEACSWKKV 470
+ R PDKA+DL+DEA + K+
Sbjct 457 ISGRFLPDKAIDLVDEAAAKLKM 479
Score = 235 bits (600), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 105/192 (54%), Positives = 148/192 (77%), Gaps = 0/192 (0%)
Query 524 DVAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPK 583
D+A I+ WTGIP+ K+ + E ++L L + L RV+GQ AV AVA+A+ RAGLS
Sbjct 615 DIAEIVSKWTGIPVSKLQQSERDKLLHLEEELHKRVVGQNPAVTAVAEAIQRSRAGLSDP 674
Query 584 NKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNLIRLDMVEYQEAHSISRLIGPPPGYM 643
+P+ +FMF+G +GVGKTELAKA+A MF++E+ L+R+DM EY E H++SRLIG PPGY+
Sbjct 675 GRPIASFMFMGPTGVGKTELAKALASYMFNTEEALVRIDMSEYMEKHAVSRLIGAPPGYV 734
Query 644 GNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCLI 703
G +EGGQLTE VR++P+SV+LFDE+E AH ++++V L +LD+G ++DS+G V FTN +I
Sbjct 735 GYEEGGQLTETVRRRPYSVILFDEIEKAHGDVFNVFLQILDDGRVTDSQGRTVSFTNTVI 794
Query 704 ILTSNIGRQHIL 715
I+TSN+G Q IL
Sbjct 795 IMTSNVGSQFIL 806
> SPBC4F6.17c
Length=803
Score = 253 bits (647), Expect = 8e-67, Method: Compositional matrix adjust.
Identities = 133/293 (45%), Positives = 190/293 (64%), Gaps = 10/293 (3%)
Query 228 ITQYGEDLTEKAAKGKLQPVIGRDEEIDRIAQILSRMMTKAPLLIGEPGVGKTAVIEGLA 287
+ +YG DLT A +GKL PVIGR+EEI R QILSR P L+G GVGKTA++EGLA
Sbjct 94 LEEYGTDLTALAKQGKLDPVIGREEEIQRTIQILSRRTKNNPALVGPAGVGKTAIMEGLA 153
Query 288 QRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEI 347
RI+ G VP+ ++ +R+I LDL L +G RG+FE+R+K +++ L+ + VILFVDE+
Sbjct 154 SRIIRGEVPESMKDKRVIVLDLGALISGAKFRGDFEERLKSVLSDLEGAEGKVILFVDEM 213
Query 348 HTLVGAGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEA 407
H L+G GK GS DA+ +LK LARG++ GATTLEEY+ +IEKDAA RRFQ ++V
Sbjct 214 HLLLGFGKAEGSIDASNLLKPALARGKLHCCGATTLEEYRKYIEKDAALARRFQAVMVNE 273
Query 408 PSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQRSFPDKALDLLDEACSW 467
PS +SIL+ +K YE H V ++D+ L S +Y+ R PDKA+DL+DEACS
Sbjct 274 PSVADTISILRGLKERYEVHHGVRITDDALVTAATYSARYITDRFLPDKAIDLVDEACSS 333
Query 468 KKVSH----------NKQIVVLKRKIEEAGKKKDETAAEELQKWKDELAALEE 510
++ ++QI+ ++ ++E K+ D T+ E +K + +L L+E
Sbjct 334 LRLQQESKPDELRRLDRQIMTIQIELESLRKETDTTSVERREKLESKLTDLKE 386
Score = 216 bits (551), Expect = 1e-55, Method: Compositional matrix adjust.
Identities = 96/192 (50%), Positives = 146/192 (76%), Gaps = 0/192 (0%)
Query 519 ALSLHDVAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRA 578
+++ D+A ++ TGIP + E ++L + + ++IGQ +A+KA+ADA+ + RA
Sbjct 464 SVTSDDIAVVVSRATGIPTTNLMRGERDKLLNMEQTIGKKIIGQDEALKAIADAVRLSRA 523
Query 579 GLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNLIRLDMVEYQEAHSISRLIGP 638
GL N+PL +F+FLG +GVGKT L KA+AE +FD++K +IR DM E+QE H+I+RLIG
Sbjct 524 GLQNTNRPLASFLFLGPTGVGKTALTKALAEFLFDTDKAMIRFDMSEFQEKHTIARLIGS 583
Query 639 PPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDF 698
PPGY+G +E G+LTEAVR+KP++V+LFDE+E AH ++ ++LL +LDEG L+DS+G +VDF
Sbjct 584 PPGYIGYEESGELTEAVRRKPYAVLLFDELEKAHHDITNLLLQVLDEGFLTDSQGRKVDF 643
Query 699 TNCLIILTSNIG 710
+ LI++TSN+G
Sbjct 644 RSTLIVMTSNLG 655
> SPBC16D10.08c
Length=905
Score = 240 bits (612), Expect = 9e-63, Method: Compositional matrix adjust.
Identities = 164/449 (36%), Positives = 249/449 (55%), Gaps = 25/449 (5%)
Query 81 TLAWHLHHDRLCLAHLLMVLLEGSRLDTP---REVITKCHGDFSHVRGAVKAVLVRLP-R 136
++A H +L H+ LL S + R ++ K GD +V + LVRLP +
Sbjct 19 SIAQSYGHSQLTPIHIAAALLSDSDSNGTTLLRTIVDKAGGDGQKFERSVTSRLVRLPAQ 78
Query 137 TPVTVPLGGAKVTPAVTEVFLRARRIAEAEGSLTGVQHVFLALLYCP-TFYKILVNSGCD 195
P P ++P ++ A + + + Q F+A+ T +L +G
Sbjct 79 DP---PPEQVTLSPESAKLLRNAHELQKTQKDSYIAQDHFIAVFTKDDTLKSLLAEAGVT 135
Query 196 VELFKEELSLAYKNTPMPNTSGTTEQLTKDFD-ITQYGEDLTEKAAKGKLQPVIGRDEEI 254
+ F E ++ N N ++ + FD + ++ DLTE A G+L PVIGR++EI
Sbjct 136 PKAF--EFAV---NNVRGNKRIDSKNAEEGFDALNKFTVDLTELARNGQLDPVIGREDEI 190
Query 255 DRIAQILSRMMTKAPLLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSA 314
R ++LSR P+LIGEPGVGKT++ EGLA+RI++ VP L +++SLD+ +L A
Sbjct 191 RRTIRVLSRRTKNNPVLIGEPGVGKTSIAEGLARRIIDDDVPANLSNCKLLSLDVGSLVA 250
Query 315 GTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSS-DATQILKVPLARG 373
G+ RGEFE+R+K ++ ++ + +ILFVDE+H L+GAG DA +LK LARG
Sbjct 251 GSKFRGEFEERIKSVLKEVEESETPIILFVDEMHLLMGAGSGGEGGMDAANLLKPMLARG 310
Query 374 EIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFHSVDLS 433
++ +GATTL EYK +IEKDAAF RRFQ I+V+ PS E +SIL+ +K YE H V +S
Sbjct 311 KLHCIGATTLAEYKKYIEKDAAFERRFQIILVKEPSIEDTISILRGLKEKYEVHHGVTIS 370
Query 434 DEVLEGIVALSDQYVKQRSFPDKALDLLDEACSWKKVSHNKQIVV---LKRKIEE----- 485
D L L+ +Y+ R PD A+DL+DEA + +V+ Q V L+RK+ +
Sbjct 371 DRALVTAAHLASRYLTSRRLPDSAIDLVDEAAAAVRVTRESQPEVLDNLERKLRQLRVEI 430
Query 486 --AGKKKDETAAEELQKWKDELAALEELT 512
++KDE + E L+ + E +EE T
Sbjct 431 RALEREKDEASKERLKAARKEAEQVEEET 459
Score = 210 bits (535), Expect = 8e-54, Method: Compositional matrix adjust.
Identities = 109/256 (42%), Positives = 160/256 (62%), Gaps = 20/256 (7%)
Query 479 LKRKIEEAGKKKDETAAEEL-----------------QKWKDELAALEELTANGRRLALS 521
LK K E+A ++ D T A +L QK K + A+ L +
Sbjct 486 LKAKAEDAERRNDFTLAADLKYYGIPDLQKRIEYLEQQKRKADAEAIANAQPGSEPLLID 545
Query 522 L---HDVAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRA 578
+ + I+ WTGIP+ ++ E R+L + +LS +VIGQ +AV AVA+A+ + RA
Sbjct 546 VVGPDQINEIVARWTGIPVTRLKTTEKERLLNMEKVLSKQVIGQNEAVTAVANAIRLSRA 605
Query 579 GLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNLIRLDMVEYQEAHSISRLIGP 638
GLS N+P+ +F+F G SG GKT L KA+A MFD E +IR+DM EY E HS+SRLIG
Sbjct 606 GLSDPNQPIASFLFCGPSGTGKTLLTKALASFMFDDENAMIRIDMSEYMEKHSVSRLIGA 665
Query 639 PPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDF 698
PPGY+G++ GGQLTE +R++P+SV+LFDE+E A + +VLL +LD+G ++ +G VD
Sbjct 666 PPGYVGHEAGGQLTEQLRRRPYSVILFDEIEKAAPEVLTVLLQVLDDGRITSGQGQVVDA 725
Query 699 TNCLIILTSNIGRQHI 714
N +II+TSN+G +++
Sbjct 726 KNAVIIMTSNLGAEYL 741
> Hs13540606
Length=707
Score = 161 bits (408), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 80/176 (45%), Positives = 119/176 (67%), Gaps = 6/176 (3%)
Query 555 LSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMF-D 613
L +IGQ+ A+ V A+ + G + PL F+FLGSSG+GKTELAK A+ M D
Sbjct 343 LKEHIIGQESAIATVGAAIRRKENGWYDEEHPL-VFLFLGSSGIGKTELAKQTAKYMHKD 401
Query 614 SEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHK 673
++K IRLDM E+QE H +++ IG PPGY+G++EGGQLT+ ++Q P++VVLFDEV+ AH
Sbjct 402 AKKGFIRLDMSEFQERHEVAKFIGSPPGYVGHEEGGQLTKKLKQCPNAVVLFDEVDKAHP 461
Query 674 NLWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGR----QHILEGYKEARALS 725
++ +++L + DEG L+D KG +D + + I+TSN+ QH L+ +EA +S
Sbjct 462 DVLTIMLQLFDEGRLTDGKGKTIDCKDAIFIMTSNVASDEIAQHALQLRQEALEMS 517
> At3g45450
Length=341
Score = 123 bits (309), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 73/177 (41%), Positives = 102/177 (57%), Gaps = 18/177 (10%)
Query 241 KGKLQPVIGRDEEIDRIAQILSRMMTKA-PLLIGEPGVGKTAVIEGLAQRIVEGAVPKGL 299
+GKL PV+GR +I R+ QIL+R + LIG+PGVGK A+ EG+AQRI G VP+ +
Sbjct 150 RGKLDPVVGRQPQIKRVVQILARRTCRNNACLIGKPGVGKRAIAEGIAQRIASGDVPETI 209
Query 300 RGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGS 359
+G+ +N+ AG E R + I + + D+ILF+DE+H L+GAG G+
Sbjct 210 KGK-------MNV-AGNCGWNEIRWRSRGKIEEVYGQSDDIILFIDEMHLLIGAGAVEGA 261
Query 360 SDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSI 416
DA ILK L R E+ +Y+ HIE D A RRFQ + V P+ E A+ I
Sbjct 262 IDAANILKPALERCEL---------QYRKHIENDPALERRFQPVKVPEPTVEEAIQI 309
> At2g25030
Length=265
Score = 100 bits (249), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 51/144 (35%), Positives = 81/144 (56%), Gaps = 33/144 (22%)
Query 551 LADILSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEE 610
L IL R+I Q V++VADA+ +AG+S N+ + +FMF+G V
Sbjct 2 LEQILHERIIAQDLDVESVADAIRCSKAGISDPNRLIASFMFMGQPSV------------ 49
Query 611 MFDSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVEN 670
+S+L+G PGY+G +GG+LTE VR++P+SVV FDE+E
Sbjct 50 ---------------------VSQLVGASPGYVGYGDGGKLTEVVRRRPYSVVQFDEIEK 88
Query 671 AHKNLWSVLLPMLDEGHLSDSKGN 694
H +++S+LL +LD+G +++S G+
Sbjct 89 PHLDVFSILLQLLDDGRITNSHGS 112
> At2g25040
Length=135
Score = 91.3 bits (225), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 40/63 (63%), Positives = 52/63 (82%), Gaps = 0/63 (0%)
Query 231 YGEDLTEKAAKGKLQPVIGRDEEIDRIAQILSRMMTKAPLLIGEPGVGKTAVIEGLAQRI 290
YG DLT+ A +GKL P+IGRD+E++R QIL RM P++IGEPGVGKTA++EGLA+RI
Sbjct 72 YGSDLTKMARQGKLPPLIGRDDEVNRCIQILCRMTESNPVIIGEPGVGKTAIVEGLAERI 131
Query 291 VEG 293
V+G
Sbjct 132 VKG 134
> At1g07200
Length=979
Score = 57.0 bits (136), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 52/191 (27%), Positives = 90/191 (47%), Gaps = 15/191 (7%)
Query 551 LADILSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFM-FLGSSGVGKTELAKAVAE 609
L +ILS +V Q +AV A++ + + + +N+ G ++ LG VGK ++A ++E
Sbjct 621 LREILSRKVAWQTEAVNAISQIICGCKTDSTRRNQASGIWLALLGPDKVGKKKVAMTLSE 680
Query 610 EMFDSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVE 669
F + N I +D E S+ + G +T + +KPHSVVL + VE
Sbjct 681 VFFGGKVNYICVDF--GAEHCSLDD------KFRGKTVVDYVTGELSRKPHSVVLLENVE 732
Query 670 NAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGR----QHILEGYK--EARA 723
A L + G + D G + N ++++TS I + H+++ K E +
Sbjct 733 KAEFPDQMRLSEAVSTGKIRDLHGRVISMKNVIVVVTSGIAKDNATDHVIKPVKFPEEQV 792
Query 724 LSASNEKAETK 734
LSA + K + K
Sbjct 793 LSARSWKLQIK 803
> YNL218w
Length=587
Score = 48.5 bits (114), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 38/115 (33%), Positives = 56/115 (48%), Gaps = 16/115 (13%)
Query 270 LLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTS-MRGEFEKRMKE 328
+L G PGVGKT++ L + + + G R ++ A T +RG FEK KE
Sbjct 174 ILWGPPGVGKTSLARLLTKTATTSSNESNV-GSRYFMIETSATKANTQELRGIFEKSKKE 232
Query 329 IITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVP-LARGEIVLVGATT 382
Q K+ +LF+DEIH + Q L +P + G+I+L+GATT
Sbjct 233 ----YQLTKRRTVLFIDEIHRF---------NKVQQDLLLPHVENGDIILIGATT 274
> At2g40130
Length=907
Score = 48.1 bits (113), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 41/185 (22%), Positives = 78/185 (42%), Gaps = 10/185 (5%)
Query 550 RLADILSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAE 609
RL D++S GQ +A + ++ AL+ ++ ++ L +G VGK ++ +AE
Sbjct 540 RLTDMVS----GQDEAARVISCALSQPPKSVTRRDVWLN---LVGPDTVGKRRMSLVLAE 592
Query 610 EMFDSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVE 669
++ SE + +D+ ++ P G + E + + P VV + +E
Sbjct 593 IVYQSEHRFMAVDLGAAEQGMGG---CDDPMRLRGKTMVDHIFEVMCRNPFCVVFLENIE 649
Query 670 NAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGRQHILEGYKEARALSASNE 729
A + L L ++ G DS G V N + ++TS+ Y E + L
Sbjct 650 KADEKLQMSLSKAIETGKFMDSHGREVGIGNTIFVMTSSSQGSATTTSYSEEKLLRVKGR 709
Query 730 KAETK 734
+ E +
Sbjct 710 QVEIR 714
> At4g30350
Length=924
Score = 47.0 bits (110), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 43/154 (27%), Positives = 74/154 (48%), Gaps = 10/154 (6%)
Query 555 LSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDS 614
L+ V Q A +VA A+ + G + K+K MF G GK+++A A+++ + S
Sbjct 577 LAKSVWWQHDAASSVAAAITECKHG-NGKSKGDIWLMFTGPDRAGKSKMASALSDLVSGS 635
Query 615 EKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKN 674
+ I L S SR+ G + EAVR+ P +V++ ++++ A
Sbjct 636 QPITISLG--------SSSRM-DDGLNIRGKTALDRFAEAVRRNPFAVIVLEDIDEADIL 686
Query 675 LWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSN 708
L + + ++ G + DS G V N +IILT+N
Sbjct 687 LRNNVKIAIERGRICDSYGREVSLGNVIIILTAN 720
> At1g50140
Length=640
Score = 45.8 bits (107), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 49/163 (30%), Positives = 71/163 (43%), Gaps = 29/163 (17%)
Query 267 KAPLLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRM 326
K LL G PG GKT + + LA G IS+ L+ + G+ EK
Sbjct 387 KGILLFGPPGTGKTLLAKALATEA----------GANFISITGSTLT--SKWFGDAEKLT 434
Query 327 KEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLARG----------EIV 376
K + +F + VI+FVDEI +L+GA S +AT+ ++ I+
Sbjct 435 KALFSFATKLAP-VIIFVDEIDSLLGARGGSSEHEATRRMRNEFMAAWDGLRSKDSQRIL 493
Query 377 LVGATTLEEYKLHIEKDAAFCRRF-QNIVVEAPSKERALSILK 418
++GAT + D A RR + I V+ P E L ILK
Sbjct 494 ILGATNRP-----FDLDDAVIRRLPRRIYVDLPDAENRLKILK 531
> 7290407
Length=479
Score = 45.1 bits (105), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 51/188 (27%), Positives = 84/188 (44%), Gaps = 29/188 (15%)
Query 270 LLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAG---TSMRGEFEKRM 326
LL G PG GKT + + L +G V N++A + RGE EK +
Sbjct 243 LLHGPPGSGKTLLAKALYSE-TQGQV------------TFFNITASIMVSKWRGESEKIL 289
Query 327 KEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLAR----GEIVLVGATT 382
+ + +R VI F DEI +L + ++++ K L + E L G
Sbjct 290 RVLFHMAAKRAPSVIFF-DEIESLTSKRDRATDHESSKRFKNELLQLLDGMEHSLNGVFV 348
Query 383 LEEYKLHIEKDAAFCRRFQ-NIVVEAP-SKERALSILKKVKTNYEKFHSVDLSDEVLEGI 440
L L + D AF RRF+ ++V+ P + ER+ I + + + S+ L+ +LE +
Sbjct 349 LASTNLPWDIDEAFLRRFEKKLLVQLPNAAERSCLINRLLGS------SISLNPRLLEQL 402
Query 441 VALSDQYV 448
V +SD +
Sbjct 403 VEISDHFT 410
> At5g40090
Length=459
Score = 45.1 bits (105), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 35/126 (27%), Positives = 63/126 (50%), Gaps = 10/126 (7%)
Query 556 SSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSE 615
S+ ++ + +K V D LA++ NK + T GS+GVGKT LA+ + E+F +
Sbjct 178 SNALVAMDRHMKVVYDLLALE------VNKEVRTIGIWGSAGVGKTTLARYIYAEIFVNF 231
Query 616 KNLIRLDMVEYQEAHSISRLIG---PPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAH 672
+ + LD VE + + + G P +G ++TEA R+ +++ D+V N
Sbjct 232 QTHVFLDNVENMK-DKLLKFEGEEDPTVIISSYHDGHEITEARRKHRKILLIADDVNNME 290
Query 673 KNLWSV 678
+ W +
Sbjct 291 QGKWII 296
> At1g64110
Length=821
Score = 44.7 bits (104), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 44/159 (27%), Positives = 72/159 (45%), Gaps = 27/159 (16%)
Query 270 LLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEI 329
LL G PG GKT + + +A+ G I++ + ++ + GE EK ++ +
Sbjct 551 LLFGPPGTGKTMLAKAIAKEA----------GASFINVSMSTIT--SKWFGEDEKNVRAL 598
Query 330 ITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLAR---------GEIVLVGA 380
T L + I+FVDE+ +++G G +A + +K GE +LV A
Sbjct 599 FT-LASKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSHWDGLMTKPGERILVLA 657
Query 381 TTLEEYKLHIEKDAAFCRRFQ-NIVVEAPSKERALSILK 418
T + L D A RRF+ I+V P+ E IL+
Sbjct 658 ATNRPFDL----DEAIIRRFERRIMVGLPAVENREKILR 692
> At4g28000
Length=726
Score = 44.3 bits (103), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 49/188 (26%), Positives = 84/188 (44%), Gaps = 33/188 (17%)
Query 270 LLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEI 329
LL G PG GKT + + +A G I++ + ++ + GE EK ++ +
Sbjct 452 LLFGPPGTGKTMMAKAIANEA----------GASFINVSMSTIT--SKWFGEDEKNVRAL 499
Query 330 ITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLAR---------GEIVLVGA 380
T L + I+FVDE+ +++G G +A + +K G+ +LV A
Sbjct 500 FT-LAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLMSNAGDRILVLA 558
Query 381 TTLEEYKLHIEKDAAFCRRFQ-NIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEG 439
T + L D A RRF+ I+V PS E IL+ + + EK ++D +
Sbjct 559 ATNRPFDL----DEAIIRRFERRIMVGLPSVESREKILRTLLSK-EKTENLDFQE----- 608
Query 440 IVALSDQY 447
+ ++D Y
Sbjct 609 LAQMTDGY 616
> At4g02470
Length=371
Score = 43.9 bits (102), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 51/192 (26%), Positives = 80/192 (41%), Gaps = 32/192 (16%)
Query 266 TKAPLLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKR 325
TK LL G PG GKT + + +A G I++ + +++ + GE EK
Sbjct 104 TKGILLFGPPGTGKTMLAKAVATEA----------GANFINISMSSIT--SKWFGEGEKY 151
Query 326 MKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLARG---------EIV 376
+K + + + VI FVDE+ +++G + G +A + +K E V
Sbjct 152 VKAVFSLASKIAPSVI-FVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDRERV 210
Query 377 LVGATTLEEYKLHIEKDAAFCRRF-QNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDE 435
LV A T + L D A RR + ++V P IL + E VD
Sbjct 211 LVLAATNRPFDL----DEAVIRRLPRRLMVNLPDATNRSKILSVILAKEEIAPDVD---- 262
Query 436 VLEGIVALSDQY 447
LE I ++D Y
Sbjct 263 -LEAIANMTDGY 273
Score = 32.0 bits (71), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 41/131 (31%), Positives = 55/131 (41%), Gaps = 29/131 (22%)
Query 505 LAALEELTANGRRLALSLHDVAHILHLWTGIPLGKMTEDE-----ISRVLRLADI-LSSR 558
L L ++ + L SL DV +TE+E +S V+ +DI +S
Sbjct 24 LQTLHDIQNENKSLKKSLKDV--------------VTENEFEKKLLSDVIPPSDIGVSFD 69
Query 559 VIGQQQAVKAVADALA---IQRAGLSPK---NKPLGTFMFLGSSGVGKTELAKAVAEEMF 612
IG + VK L +QR L K KP + G G GKT LAKAVA E
Sbjct 70 DIGALENVKETLKELVMLPLQRPELFDKGQLTKPTKGILLFGPPGTGKTMLAKAVATE-- 127
Query 613 DSEKNLIRLDM 623
+ N I + M
Sbjct 128 -AGANFINISM 137
> At3g19740
Length=439
Score = 43.5 bits (101), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 47/163 (28%), Positives = 71/163 (43%), Gaps = 29/163 (17%)
Query 267 KAPLLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRM 326
K LL G PG GKT + + LA G IS+ L+ + G+ EK
Sbjct 186 KGILLFGPPGTGKTLLAKALATEA----------GANFISITGSTLT--SKWFGDAEKLT 233
Query 327 KEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLARG----------EIV 376
K + +F + VI+FVDE+ +L+GA + +AT+ ++ I+
Sbjct 234 KALFSFASKLAP-VIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKDSQRIL 292
Query 377 LVGATTLEEYKLHIEKDAAFCRRF-QNIVVEAPSKERALSILK 418
++GAT + D A RR + I V+ P E L ILK
Sbjct 293 ILGATN-----RPFDLDDAVIRRLPRRIYVDLPDAENRLKILK 330
> At3g27120
Length=327
Score = 43.1 bits (100), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 67/253 (26%), Positives = 105/253 (41%), Gaps = 50/253 (19%)
Query 267 KAPLLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMR-GEFEKR 325
K LL G PG GKT + + +A G + ++ S+ TS GE EK
Sbjct 84 KGLLLFGPPGTGKTMIGKAIA-------------GEAKATFFYISASSLTSKWIGEGEKL 130
Query 326 MKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPL---------ARGEIV 376
++ + R+ VI FVDEI +L+ K+ G ++++ LK +I+
Sbjct 131 VRALFGVASCRQPAVI-FVDEIDSLLSQRKSDGEHESSRRLKTQFLIEMEGFDSGSEQIL 189
Query 377 LVGATTLEEYKLHIEKDAAFCRRF-QNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDE 435
L+GAT + E D A RR + + + PS E I++ + +K LSD+
Sbjct 190 LIGATNRPQ-----ELDEAARRRLTKRLYIPLPSSEARAWIIQNL---LKKDGLFTLSDD 241
Query 436 VLEGIVALSDQYV---KQRSFPDKALDLLDEACSWKKVSHNKQIVVLKRKIEEAGKKKDE 492
+ I L++ Y + D + L EA LKR I+ KD+
Sbjct 242 DMNIICNLTEGYSGSDMKNLVKDATMGPLREA--------------LKRGIDITNLTKDD 287
Query 493 TAAEELQKWKDEL 505
LQ +KD L
Sbjct 288 MRLVTLQDFKDAL 300
> 7290516
Length=339
Score = 42.7 bits (99), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 40/176 (22%), Positives = 71/176 (40%), Gaps = 23/176 (13%)
Query 546 SRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAK 605
+R+ L L +IGQ + + AL A + KPL F G G GK +A+
Sbjct 60 ARIDELERSLERTLIGQHIVRQHIVPALKAHIASGNKSRKPL-VISFHGQPGTGKNFVAE 118
Query 606 AVAEEMFDSEKNLIRLDMVEYQEAHSISRLIG----PPPGYMGNDE---GGQLTEAVRQK 658
+A+ M+ ++ ++ +++ +G P + N + + +R
Sbjct 119 QIADAMY-----------LKGSRSNYVTKYLGQADFPKESEVSNYRVKINNAVRDTLRSC 167
Query 659 PHSVVLFDEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGRQHI 714
P S+ +FDEV+ ++ L ++D D N T + I SN HI
Sbjct 168 PRSLFIFDEVDKMPSGVFDQLTSLVDYNAFVDGTDN----TKAIFIFLSNTAGSHI 219
> At1g02890_2
Length=563
Score = 42.0 bits (97), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 52/192 (27%), Positives = 81/192 (42%), Gaps = 36/192 (18%)
Query 266 TKAPLLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKR 325
TK LL G PG GKT + + +A GA + +N+S +S+ + EK
Sbjct 300 TKGILLFGPPGTGKTMLAKAVATE--AGA-------------NFINISM-SSITSKGEKY 343
Query 326 MKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILK---------VPLARGEIV 376
+K + + + VI FVDE+ +++G + G +A + +K + E V
Sbjct 344 VKAVFSLASKIAPSVI-FVDEVDSMLGRRENPGEHEAMRKMKNEFMINWDGLRTKDKERV 402
Query 377 LVGATTLEEYKLHIEKDAAFCRRF-QNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDE 435
LV A T + L D A RR + ++V P IL + E VD
Sbjct 403 LVLAATNRPFDL----DEAVIRRLPRRLMVNLPDSANRSKILSVILAKEEMAEDVD---- 454
Query 436 VLEGIVALSDQY 447
LE I ++D Y
Sbjct 455 -LEAIANMTDGY 465
Score = 32.7 bits (73), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 33/97 (34%), Positives = 45/97 (46%), Gaps = 17/97 (17%)
Query 540 MTEDEISRVLRLADILSSRVIGQQ----QAVKAVADAL------AIQRAGLSPK---NKP 586
+TE+E + L L+D++ IG A++ V D L +QR L K KP
Sbjct 241 VTENEFEKKL-LSDVIPPSDIGVSFSDIGALENVKDTLKELVMLPLQRPELFGKGQLTKP 299
Query 587 LGTFMFLGSSGVGKTELAKAVAEEMFDSEKNLIRLDM 623
+ G G GKT LAKAVA E + N I + M
Sbjct 300 TKGILLFGPPGTGKTMLAKAVATE---AGANFINISM 333
> CE28132
Length=813
Score = 42.0 bits (97), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 48/166 (28%), Positives = 72/166 (43%), Gaps = 32/166 (19%)
Query 270 LLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEI 329
LL G PG GKT + + +A G+ + +LLN+ G S R +
Sbjct 572 LLCGPPGCGKTLLAKAVANET-------GMNFISVKGPELLNMYVGESERA--------V 616
Query 330 ITFLQRRK--KDVILFVDEIHTLV---GAGKTSGSSDATQILKVPL----ARGEIVLVGA 380
T QR + + ++F DEI LV G++SG + L + R ++ L+GA
Sbjct 617 RTVFQRARDSQPCVIFFDEIDALVPKRSHGESSGGARLVNQLLTEMDGVEGRQKVFLIGA 676
Query 381 TTLEEYKLHIEKDAAFCR--RFQNIV-VEAPSKERALSILKKVKTN 423
T + DAA R R I+ V+ PS E + IL+K N
Sbjct 677 TNRPDI-----VDAAILRPGRLDKILFVDFPSVEDRVDILRKSTKN 717
> At5g57710
Length=990
Score = 41.6 bits (96), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 25/119 (21%), Positives = 57/119 (47%), Gaps = 8/119 (6%)
Query 590 FMFLGSSGVGKTELAKAVAEEMFDSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGG 649
+F G VGK ++ A++ ++ + N I + + Q+A + + G
Sbjct 655 LLFSGPDRVGKRKMVSALSSLVYGT--NPIMIQLGSRQDAGDGNS------SFRGKTALD 706
Query 650 QLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSN 708
++ E V++ P SV+L ++++ A + + +D G + DS G + N + ++T++
Sbjct 707 KIAETVKRSPFSVILLEDIDEADMLVRGSIKQAMDRGRIRDSHGREISLGNVIFVMTAS 765
Score = 34.7 bits (78), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 57/295 (19%), Positives = 125/295 (42%), Gaps = 61/295 (20%)
Query 172 VQHVFLALLYCPTFYKILVNSGCDVELFKEELSLAYKN----TPMPNTS--------GTT 219
++ + +++L P+ +++ + K + + N TP+P+ S G
Sbjct 130 LEQLIISILDDPSVSRVMREASFSSPAVKATIEQSLNNSVTPTPIPSVSSVGLNFRPGGG 189
Query 220 EQLTKDFDITQYGEDLTEKAAKGKLQPVIGRDEEIDRIAQILSRMMTKAPLLIGEPGVGK 279
+T++ + L + A+ +Q + ++++++R+ IL R K P+L+G+ G+
Sbjct 190 GPMTRN---SYLNPRLQQNASS--VQSGVSKNDDVERVMDILGRAKKKNPVLVGDSEPGR 244
Query 280 TAVIEGLAQRIVEGAVPK-GLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKK 338
VI + ++I G V ++ +++SL+ +S+ ++ R+KE+ LQ R K
Sbjct 245 --VIREILKKIEVGEVGNLAVKNSKVVSLE--EISSDKAL------RIKELDGLLQTRLK 294
Query 339 D--------VILFVDEIHTLVGAGKTSGSSDATQILKVPLAR--------------GEIV 376
+ VIL + ++ LV + S+ + V + R G +
Sbjct 295 NSDPIGGGGVILDLGDLKWLV---EQPSSTQPPATVAVEIGRTAVVELRRLLEKFEGRLW 351
Query 377 LVGATTLEEY----KLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKF 427
+G T E Y H + + + ++ +AP A + ++ N E F
Sbjct 352 FIGTATCETYLRCQVYHPSVETDWDLQAVSVAAKAP----ASGVFPRLANNLESF 402
> At5g45720
Length=900
Score = 41.2 bits (95), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 31/132 (23%), Positives = 61/132 (46%), Gaps = 14/132 (10%)
Query 559 VIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMF-----D 613
++GQ V+A+++A+A +R GL ++F G +G GKT A+ A +
Sbjct 357 LLGQNLVVQALSNAIAKRRVGL--------LYVFHGPNGTGKTSCARVFARALNCHSTEQ 408
Query 614 SEKNLIRLDMVEYQEAHS-ISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAH 672
S+ + V Y + + R +GP + + + +QK V++FD+ +
Sbjct 409 SKPCGVCSSCVSYDDGKNRYIREMGPVKSFDFENLLDKTNIRQQQKQQLVLIFDDCDTMS 468
Query 673 KNLWSVLLPMLD 684
+ W+ L ++D
Sbjct 469 TDCWNTLSKIVD 480
> Hs14150179
Length=939
Score = 40.8 bits (94), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 42/157 (26%), Positives = 63/157 (40%), Gaps = 19/157 (12%)
Query 270 LLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEI 329
L G PG GKT V LA +G +R+ + GE E++++ +
Sbjct 464 LFYGPPGTGKTLVARALANECSQG-------DKRVAFFMRKGADCLSKWVGESERQLRLL 516
Query 330 ITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLA-------RGEIVLVGATT 382
+ + +I F DEI L + + I+ LA RGEIV++GAT
Sbjct 517 FDQAYQMRPSIIFF-DEIDGLAPVRSSRQDQIHSSIVSTLLALMDGLDSRGEIVVIGATN 575
Query 383 -LEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILK 418
L+ + + F R F + P KE ILK
Sbjct 576 RLDSIDPALRRPGRFDREF---LFSLPDKEARKEILK 609
Score = 33.1 bits (74), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 30/117 (25%), Positives = 53/117 (45%), Gaps = 27/117 (23%)
Query 583 KNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNLIRLDMVEYQEAHSISRLIGPPPGY 642
K +P +F G G GKT +A+A+A E +K R+ + A +S+ +G
Sbjct 456 KIQPPRGCLFYGPPGTGKTLVARALANECSQGDK---RVAFFMRKGADCLSKWVG----- 507
Query 643 MGNDEGGQL----TEAVRQKPHSVVLFDEV-----------ENAHKNLWSVLLPMLD 684
+ QL +A + +P S++ FDE+ + H ++ S LL ++D
Sbjct 508 ---ESERQLRLLFDQAYQMRP-SIIFFDEIDGLAPVRSSRQDQIHSSIVSTLLALMD 560
> 7297651
Length=369
Score = 40.4 bits (93), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 46/178 (25%), Positives = 77/178 (43%), Gaps = 30/178 (16%)
Query 267 KAPLLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRM 326
K LL G PG GKT + + A+ G R I+LD+ L+ GE +K
Sbjct 133 KGVLLHGPPGCGKTLIAKATAKEA----------GMRFINLDVAILT--DKWYGESQKLT 180
Query 327 KEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPL----------ARGEIV 376
+ + L R + I+F+DEI + + + + +AT ++K A ++
Sbjct 181 SAVFS-LASRIEPCIIFIDEIDSFLRS-RNMNDHEATAMMKTQFMMLWDGLSTNANSTVI 238
Query 377 LVGATTLEEYKLHIEKDAAFCRRF-QNIVVEAPSKERALSILKKVKTNYEKFHSVDLS 433
++GAT + + D A RR + PS+ + ILK + + E VDL+
Sbjct 239 VMGATNRPQ-----DLDKAIVRRMPAQFHIGLPSETQRKDILKLILQSEEVSQDVDLN 291
> YGR028w
Length=362
Score = 40.0 bits (92), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 51/207 (24%), Positives = 89/207 (42%), Gaps = 44/207 (21%)
Query 270 LLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEI 329
LL G PG GKT + + LA+ GA +R I+ GE K + +
Sbjct 130 LLYGPPGCGKTMLAKALAKE--SGANFISIRMSSIMD----------KWYGESNKIVDAM 177
Query 330 ITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVP--------LARGEIVLVGAT 381
+ L + + I+F+DEI + + ++S + T LK L G ++++GAT
Sbjct 178 FS-LANKLQPCIIFIDEIDSFLRE-RSSTDHEVTATLKAEFMTLWDGLLNNGRVMIIGAT 235
Query 382 TLEEYKLHIEKDAAFCRRF-QNIVVEAPSKE---RALSILKK-------------VKTNY 424
+++ + D AF RR + +V P + + LS+L K + N
Sbjct 236 N----RIN-DIDDAFLRRLPKRFLVSLPGSDQRYKILSVLLKDTKLDEDEFDLQLIADNT 290
Query 425 EKFHSVDLSDEVLEGIVALSDQYVKQR 451
+ F DL + E + + +Y+KQ+
Sbjct 291 KGFSGSDLKELCREAALDAAKEYIKQK 317
> YPR173c
Length=437
Score = 40.0 bits (92), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 43/150 (28%), Positives = 71/150 (47%), Gaps = 30/150 (20%)
Query 262 SRMMTKAPLLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGE 321
+R T LL G PG GK+ + + +A + S DL+ + GE
Sbjct 162 NRKPTSGILLYGPPGTGKSYLAKAVATEANSTFF-------SVSSSDLV-----SKWMGE 209
Query 322 FEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPL----------A 371
EK +K++ + R K I+F+DE+ L G + G S+A++ +K L +
Sbjct 210 SEKLVKQLFA-MARENKPSIIFIDEVDALTGT-RGEGESEASRRIKTELLVQMNGVGNDS 267
Query 372 RGEIVLVGATTLEEYKLHIEKDAAFCRRFQ 401
+G +VL GAT + ++L D+A RRF+
Sbjct 268 QGVLVL-GATNI-PWQL----DSAIRRRFE 291
> Hs18426902
Length=665
Score = 39.3 bits (90), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 56/205 (27%), Positives = 92/205 (44%), Gaps = 27/205 (13%)
Query 232 GEDLTEKAAKGKLQPVIGRDEEIDRIAQILSRMMT-KAPLLI--GEPGVGKTAVIEGLAQ 288
G+ L + LQ G+ + + + + S + T + P LI G PG GKT + +A
Sbjct 224 GKPLADTMRPDTLQDYFGQSKAVGQDTLLRSLLETNEIPSLILWGPPGCGKTTLAHIIAS 283
Query 289 RIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIH 348
+ ++ R ++L N A T+ + K+ + +F +R+ ILF+DEIH
Sbjct 284 NSKKHSI-------RFVTLSATN--AKTNDVRDVIKQAQNEKSFFKRK---TILFIDEIH 331
Query 349 TLVGAGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAP 408
+ + S T + V G I L+GATT E + +AA R + IV+E
Sbjct 332 ------RFNKSQQDTFLPHVEC--GTITLIGATT-ENPSFQV--NAALLSRCRVIVLEKL 380
Query 409 SKERALSILKKVKTNYEKFHSVDLS 433
E ++IL + N H +D S
Sbjct 381 PVEAMVTILMRA-INSLGIHVLDSS 404
> Hs11545833
Length=674
Score = 39.3 bits (90), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 49/194 (25%), Positives = 87/194 (44%), Gaps = 30/194 (15%)
Query 263 RMMTKAPLLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEF 322
R K LL G PG GKT + + +A + G S+ +L+ + GE
Sbjct 431 RGPPKGILLFGPPGTGKTLIGKCIASQ----------SGATFFSISASSLT--SKWVGEG 478
Query 323 EKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLARGEIVLVGATT 382
EK ++ + + R ++ ++F+DEI +L+ + + G ++++ +K + L GATT
Sbjct 479 EKMVRALFA-VARCQQPAVIFIDEIDSLL-SQRGDGEHESSRRIKTEFL---VQLDGATT 533
Query 383 LEEYKLHI--------EKDAAFCRRF-QNIVVEAPSKERALSILKKVKTNYEKFHSVDLS 433
E ++ + E D A RR + + + P S K++ N LS
Sbjct 534 SSEDRILVVGATNRPQEIDEAARRRLVKRLYIPLPEA----SARKQIVINLMSKEQCCLS 589
Query 434 DEVLEGIVALSDQY 447
+E +E IV SD +
Sbjct 590 EEEIEQIVQQSDAF 603
> ECU01g1230
Length=780
Score = 38.9 bits (89), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 58/226 (25%), Positives = 96/226 (42%), Gaps = 34/226 (15%)
Query 220 EQLTKDFDITQYGEDLTEKAAKGKLQPVIGRDEEIDRIAQILSRMMTKAP---LLIGEPG 276
E++ K+F++ Y + +A K++ ++ E R +Q+ S++ K P LL G PG
Sbjct 196 EEVEKEFNMVGYDDVGGCRAQMAKIRELV---ELPLRHSQLYSKIGVKPPKGILLYGPPG 252
Query 277 VGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRR 336
GKT + +A GA + G I+S M GE E +++ ++
Sbjct 253 TGKTLIARAIANET--GAFLFLINGPEIMS----------KMAGESESNLRKAFEEAEKN 300
Query 337 KKDVILFVDEIHTLVGAGKTSGSSDATQILKVPL-------ARGEIVLVGATTLEEYKLH 389
I+F+DEI L + S +I+ L AR ++++GAT
Sbjct 301 SP-AIIFIDEIDALAPKREKSQGEVERRIVSQLLTLMDGMKARSNVIVLGATNRPN---- 355
Query 390 IEKDAAFCR--RF-QNIVVEAPSKERALSILKKVKTNYEKFHSVDL 432
D A R RF + I + P + L IL+ N + VDL
Sbjct 356 -SIDPALRRYGRFDREIEIGVPDETGRLEILRIHTKNMKMSEDVDL 400
> Hs17865802
Length=444
Score = 38.1 bits (87), Expect = 0.063, Method: Compositional matrix adjust.
Identities = 41/141 (29%), Positives = 66/141 (46%), Gaps = 27/141 (19%)
Query 270 LLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEI 329
LL G PG GK+ + + +A I S DL+ + GE EK +K +
Sbjct 171 LLFGPPGTGKSYLAKAVATEANNSTF------FSISSSDLV-----SKWLGESEKLVKNL 219
Query 330 ITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILK---------VPLARGEIVLVGA 380
L R K I+F+DEI +L G+ ++ S+A + +K V + I+++GA
Sbjct 220 FQ-LARENKPSIIFIDEIDSLCGS-RSENESEAARRIKTEFLVQMQGVGVDNDGILVLGA 277
Query 381 TTLEEYKLHIEKDAAFCRRFQ 401
T + + L D+A RRF+
Sbjct 278 TNI-PWVL----DSAIRRRFE 293
> At2g34560
Length=393
Score = 38.1 bits (87), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 44/159 (27%), Positives = 66/159 (41%), Gaps = 34/159 (21%)
Query 267 KAPLLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAG---TSMRGEFE 323
K LL G PG GKT + + +A + N+SA + RG+ E
Sbjct 146 KGILLFGPPGTGKTMLAKAVATE---------------CNTTFFNISASSVVSKWRGDSE 190
Query 324 KRMKEIITFLQRRKKDVILFVDEIHTLVG--AGKTSGSSDATQILKVPL--------ARG 373
K ++ ++ L R +F+DEI ++ G+ +A++ LK L
Sbjct 191 KLIR-VLFDLARHHAPSTIFLDEIDAIISQRGGEGRSEHEASRRLKTELLIQMDGLQKTN 249
Query 374 EIVLVGATTLEEYKLHIEKDAAFCRRFQN-IVVEAPSKE 411
E+V V A T L E DAA RR + I+V P E
Sbjct 250 ELVFVLAAT----NLPWELDAAMLRRLEKRILVPLPDPE 284
> At1g05910
Length=1069
Score = 38.1 bits (87), Expect = 0.081, Method: Compositional matrix adjust.
Identities = 35/120 (29%), Positives = 51/120 (42%), Gaps = 15/120 (12%)
Query 270 LLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEI 329
LL G PG GKT + LA + R+ D+L + GE E+++K +
Sbjct 286 LLCGPPGTGKTLIARALACAASKAGQKVSFYMRK--GADVL-----SKWVGEAERQLKLL 338
Query 330 ITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLA-------RGEIVLVGATT 382
QR + +I F DEI L + I+ LA RG++VL+GAT
Sbjct 339 FEEAQRNQPSIIFF-DEIDGLAPVRSSKQEQIHNSIVSTLLALMDGLDSRGQVVLIGATN 397
> CE28241
Length=339
Score = 37.7 bits (86), Expect = 0.091, Method: Compositional matrix adjust.
Identities = 49/191 (25%), Positives = 86/191 (45%), Gaps = 30/191 (15%)
Query 270 LLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEI 329
LL G PG GKT LA+ + A G R I+L + NL+ GE +K +
Sbjct 118 LLYGPPGCGKTL----LAKAVARAA------GCRFINLQVSNLT--DKWYGESQKLAAAV 165
Query 330 ITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPL---------ARGEIVLVGA 380
+ Q+ + I+F+DEI + + + S ++T ++K + +I+++GA
Sbjct 166 FSVAQKFQP-TIIFIDEIDSFL-RDRQSHDHESTAMMKAQFMTLWDGFSSSGDQIIVMGA 223
Query 381 TTLEEYKLHIEKDAAFCRRFQ-NIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEG 439
T + DAA RR V P+ ++ IL + N + ++V+L E+ +
Sbjct 224 TNRPR-----DVDAAILRRMTARFQVPVPNAKQRSQILNVILRNEKINNTVNLG-EIAQA 277
Query 440 IVALSDQYVKQ 450
LS +K+
Sbjct 278 AEGLSGSDLKE 288
> CE28240
Length=342
Score = 37.7 bits (86), Expect = 0.096, Method: Compositional matrix adjust.
Identities = 49/191 (25%), Positives = 86/191 (45%), Gaps = 30/191 (15%)
Query 270 LLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEI 329
LL G PG GKT LA+ + A G R I+L + NL+ GE +K +
Sbjct 121 LLYGPPGCGKTL----LAKAVARAA------GCRFINLQVSNLT--DKWYGESQKLAAAV 168
Query 330 ITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPL---------ARGEIVLVGA 380
+ Q+ + I+F+DEI + + + S ++T ++K + +I+++GA
Sbjct 169 FSVAQKFQP-TIIFIDEIDSFL-RDRQSHDHESTAMMKAQFMTLWDGFSSSGDQIIVMGA 226
Query 381 TTLEEYKLHIEKDAAFCRRFQ-NIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEG 439
T + DAA RR V P+ ++ IL + N + ++V+L E+ +
Sbjct 227 TNRPR-----DVDAAILRRMTARFQVPVPNAKQRSQILNVILRNEKINNTVNLG-EIAQA 280
Query 440 IVALSDQYVKQ 450
LS +K+
Sbjct 281 AEGLSGSDLKE 291
> Hs5901990
Length=491
Score = 37.4 bits (85), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 43/178 (24%), Positives = 71/178 (39%), Gaps = 39/178 (21%)
Query 216 SGTTEQLTKDFDITQYGEDLTEKAAKGKL--QPVIGRDEEIDRI--------AQILSRMM 265
+ TE T FD T Y +DL E + + P + D+ D + A +L M
Sbjct 172 AAVTEPETNKFDSTGYDKDLVEALERDIISQNPNVRWDDIADLVEAKKLLKEAVVLPMWM 231
Query 266 T----------KAPLLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAG 315
K L++G PG GKT + + +A N+S+
Sbjct 232 PEFFKGIRRPWKGVLMVGPPGTGKTLLAKAVATE---------------CKTTFFNVSSS 276
Query 316 T---SMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPL 370
T RGE EK ++ ++ + R +F+DEI ++ TS +A++ +K L
Sbjct 277 TLTSKYRGESEKLVR-LLFEMARFYSPATIFIDEIDSICSRRGTSEEHEASRRVKAEL 333
> At2g27600
Length=435
Score = 37.0 bits (84), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 45/155 (29%), Positives = 64/155 (41%), Gaps = 26/155 (16%)
Query 263 RMMTKAPLLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEF 322
R +A LL G PG GK+ + + +A + S DL+ + GE
Sbjct 162 RRPWRAFLLYGPPGTGKSYLAKAVATEADSTFF-------SVSSSDLV-----SKWMGES 209
Query 323 EKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPL--------ARGE 374
EK + + + R I+FVDEI +L G S+A++ +K L E
Sbjct 210 EKLVSNLFE-MARESAPSIIFVDEIDSLCGTRGEGNESEASRRIKTELLVQMQGVGHNDE 268
Query 375 IVLVGATTLEEYKLHIEKDAAFCRRF-QNIVVEAP 408
VLV A T Y L D A RRF + I + P
Sbjct 269 KVLVLAATNTPYAL----DQAIRRRFDKRIYIPLP 299
Score = 35.8 bits (81), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 32/123 (26%), Positives = 54/123 (43%), Gaps = 20/123 (16%)
Query 553 DILSSRVIGQQQAVKAVADALAIQ---RAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAE 609
+I S V G + A +A+ +A+ + + K +P F+ G G GK+ LAKAVA
Sbjct 128 NIKWSDVAGLESAKQALQEAVILPVKFPQFFTGKRRPWRAFLLYGPPGTGKSYLAKAVAT 187
Query 610 EMFDSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEG--GQLTEAVRQKPHSVVLFDE 667
E + ++ D+V +MG E L E R+ S++ DE
Sbjct 188 EADSTFFSVSSSDLVS---------------KWMGESEKLVSNLFEMARESAPSIIFVDE 232
Query 668 VEN 670
+++
Sbjct 233 IDS 235
> SPAC328.04
Length=741
Score = 37.0 bits (84), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 42/94 (44%), Gaps = 13/94 (13%)
Query 270 LLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEI 329
LL G PG GKT + +A R + + S + GE EK ++ +
Sbjct 496 LLFGPPGTGKTMLARAVATE------------SRSVFFSISASSLTSKFLGESEKLVRAL 543
Query 330 ITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDAT 363
T L ++ I+FVDEI +L+ A + G+ T
Sbjct 544 FT-LAKKLSPSIIFVDEIDSLLSARSSDGNEHET 576
> 7296674
Length=669
Score = 37.0 bits (84), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 38/164 (23%), Positives = 69/164 (42%), Gaps = 35/164 (21%)
Query 270 LLIGEPGVGKTAVIEGLAQRIVEGAVPKGLRGRRIISLDLLNLSAGT---SMRGEFEKRM 326
L++G PG GKT + + +A N+S+ T RGE EK +
Sbjct 427 LMVGPPGTGKTMLAKAVATE---------------CGTTFFNVSSSTLTSKYRGESEKLV 471
Query 327 KEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPL-----------ARGEI 375
+ ++ + R +F+DEI L + + +A++ K L ++
Sbjct 472 R-LLFEMARFYAPSTIFIDEIDALCASRGSDSEHEASRRFKAELLIQMDGLNASMQEEKV 530
Query 376 VLVGATTLEEYKLHIEKDAAFCRRFQN-IVVEAPSKERALSILK 418
++V A T + + D AF RRF+ I + P++ ++LK
Sbjct 531 IMVLAATNHPWDI----DEAFRRRFEKRIYIPLPNEGTRSALLK 570
> HsM4758682
Length=937
Score = 37.0 bits (84), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 65/265 (24%), Positives = 109/265 (41%), Gaps = 35/265 (13%)
Query 469 KVSHNKQIVVLKRKI--EEAGKKKDETAAEELQKWKDELAAL-----------EELTANG 515
K +H K ++ + KI +E G +KD+ A E +K+++ L L EEL+ G
Sbjct 366 KQTHRKYLLQEQLKIIKKELGLEKDDKDAIE-EKFRERLKELVVPKHVMDVVDEELSKLG 424
Query 516 --RRLALSLHDVAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADAL 573
+ + + L T IP GK + + + + R +L G + K + + +
Sbjct 425 LLDNHSSEFNVTRNYLDWLTSIPWGKYSNENLD-LARAQAVLEEDHYGMEDVKKRILEFI 483
Query 574 AIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNLIRLDMVEYQEAHSIS 633
A+ + S + K L F G GVGKT +A+++A + M + E
Sbjct 484 AVSQLRGSTQGKIL---CFYGPPGVGKTSIARSIARALNREYFRFSVGGMTDVAEIKGHR 540
Query 634 R-LIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLW----SVLLPMLDE--- 685
R +G PG + TE + ++L DEV+ + S LL +LD
Sbjct 541 RTYVGAMPGKIIQCLKKTKTE------NPLILIDEVDKIGRGYQGDPSSALLELLDPEQN 594
Query 686 -GHLSDSKGNRVDFTNCLIILTSNI 709
L VD + L I T+N+
Sbjct 595 ANFLDHYLDVPVDLSKVLFICTANV 619
Lambda K H
0.317 0.133 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 21294693154
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40