bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0529_orf2
Length=204
Score E
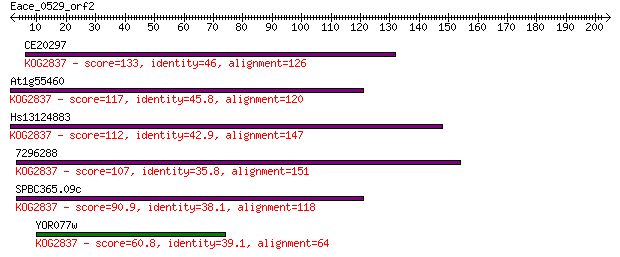
Sequences producing significant alignments: (Bits) Value
CE20297 133 2e-31
At1g55460 117 2e-26
Hs13124883 112 6e-25
7296288 107 2e-23
SPBC365.09c 90.9 1e-18
YOR077w 60.8 2e-09
> CE20297
Length=404
Score = 133 bits (335), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 58/126 (46%), Positives = 86/126 (68%), Gaps = 0/126 (0%)
Query 6 FMDEFSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEFVQYLG 65
++ +FS +FE+ FM+L+RT Y RV AN VYN + D+ H+HMNST+W +L+ FVQYLG
Sbjct 64 YLRQFSNDFEKNFMQLLRTSYGTKRVRANEVYNAFIKDKGHVHMNSTVWHSLTGFVQYLG 123
Query 66 ATQKCKIEHTPKGWYVEYIDHEELAKKEEEAKRRKIEKSQEDARMEQIQAMVEESRRRGG 125
++ KCKI+ KGWY+ YID E L +KEE+ ++++ EK E+ M+ + MV+ + G
Sbjct 124 SSGKCKIDEGDKGWYIAYIDQEALIRKEEDQRKQQQEKDDEERHMQIMDGMVQRGKELAG 183
Query 126 FQEPEY 131
E EY
Sbjct 184 DDEHEY 189
> At1g55460
Length=411
Score = 117 bits (293), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 55/122 (45%), Positives = 82/122 (67%), Gaps = 2/122 (1%)
Query 1 QRPGKFMDEFSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEF 60
Q P + +D +S EFE+ F+ LMR + +R+ A VYN + DRHH+HMNST W TL+EF
Sbjct 59 QNPTRVVDGYSEEFEQTFLDLMRRSHRFSRIAATVVYNEYINDRHHVHMNSTEWATLTEF 118
Query 61 VQYLGATQKCKIEHTPKGWYVEYIDH--EELAKKEEEAKRRKIEKSQEDARMEQIQAMVE 118
+++LG T KCK+E TPKGW++ YID E L K+ + KR K + ++E+ + +IQ +E
Sbjct 119 IKHLGKTGKCKVEETPKGWFITYIDRDSETLFKERLKNKRVKSDLAEEEKQEREIQRQIE 178
Query 119 ES 120
+
Sbjct 179 RA 180
> Hs13124883
Length=393
Score = 112 bits (280), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 63/155 (40%), Positives = 91/155 (58%), Gaps = 15/155 (9%)
Query 1 QRPGKFMDEFSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEF 60
+ P +FMD FS EF +F++L+R R+ RV N VYN + R HIHMN+T W TL++F
Sbjct 59 ENPQQFMDYFSEEFRNDFLELLRRRFGTKRVHNNIVYNEYISHREHIHMNATQWETLTDF 118
Query 61 VQYLGATQKCKIEHTPKGWYVEYIDHEELAKKEEEAKRRKIE----KSQEDARMEQIQAM 116
++LG CK++ TPKGWY++YID ++ E RR++E K Q+ E+
Sbjct 119 TKWLGREGLCKVDETPKGWYIQYID------RDPETIRRQLELEKKKKQDLDDEEKTAKF 172
Query 117 VEESRRRG--GFQE--PEYTPLQREEGHTVALTFN 147
+EE RRG G ++ P +T L RE +TFN
Sbjct 173 IEEQVRRGLEGKEQEVPTFTELSRENDEE-KVTFN 206
> 7296288
Length=390
Score = 107 bits (266), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 54/159 (33%), Positives = 96/159 (60%), Gaps = 8/159 (5%)
Query 3 PGKFMDEFSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEFVQ 62
PGKF+ FS+EF + +M+L+R R+ R AN +Y + + HIHMN+T W+TLS++V+
Sbjct 61 PGKFLHSFSKEFSDGYMELLRRRFGTKRTSANKIYQEYIAHKEHIHMNATRWLTLSDYVK 120
Query 63 YLGATQKCKIEHTPKGWYVEYIDH--EELAKKEEEAKRRKIEKSQEDARMEQIQAMVEES 120
+LG T + + T KGW+V YID E + ++ + ++ K+EK E+ + I+ ++ +
Sbjct 121 WLGRTGQVIADETEKGWFVTYIDRSPEAMERQAKADRKEKMEKDDEERMADFIEQQIKNA 180
Query 121 RRRGGFQE---PEYTPLQREEGHTVALTF---NKTQPES 153
+ + G ++ ++T L+REE + L K QP++
Sbjct 181 KAKDGEEDEGQEKFTELKREENEPLKLDIRLEKKFQPDT 219
> SPBC365.09c
Length=304
Score = 90.9 bits (224), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 45/124 (36%), Positives = 73/124 (58%), Gaps = 6/124 (4%)
Query 3 PGKFMDEFSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEFVQ 62
PGK + +FS +F +F+ L+RT + ++ N Y + D++H+HMN+T W TLSEF +
Sbjct 61 PGKRIQDFSNQFLRDFISLLRTAHGEKKIHFNQFYQEYIRDKNHVHMNATRWHTLSEFCK 120
Query 63 YLGATQKCKIEHTPKGWYVEYIDHE--ELAKKEEEAKRRKIEKSQEDARM----EQIQAM 116
+LG C++E KG+++ YID + + E KR + EKS E+ R+ EQI+
Sbjct 121 FLGRQGMCRVEENEKGFFISYIDKNPANILRNEANKKRERQEKSDEEQRLRLLDEQIKRA 180
Query 117 VEES 120
E +
Sbjct 181 YESA 184
> YOR077w
Length=232
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 25/64 (39%), Positives = 44/64 (68%), Gaps = 0/64 (0%)
Query 10 FSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEFVQYLGATQK 69
++ +FE+ F++L++ R+ + AN VYN V DR H+HMN+T+ +L++FV+YLG K
Sbjct 63 YNIQFEKGFLQLLKQRHGEKWIDANKVYNEYVQDRDHVHMNATMHRSLTQFVRYLGRAGK 122
Query 70 CKIE 73
++
Sbjct 123 VDVD 126
Lambda K H
0.314 0.127 0.354
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3622842326
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40