bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0516_orf1
Length=181
Score E
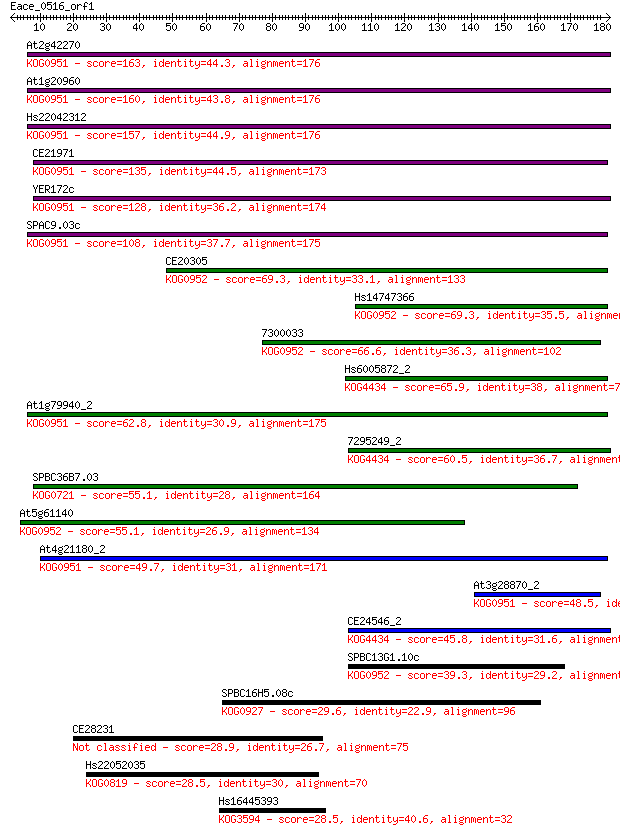
Sequences producing significant alignments: (Bits) Value
At2g42270 163 2e-40
At1g20960 160 2e-39
Hs22042312 157 1e-38
CE21971 135 5e-32
YER172c 128 7e-30
SPAC9.03c 108 5e-24
CE20305 69.3 3e-12
Hs14747366 69.3 4e-12
7300033 66.6 3e-11
Hs6005872_2 65.9 4e-11
At1g79940_2 62.8 4e-10
7295249_2 60.5 2e-09
SPBC36B7.03 55.1 8e-08
At5g61140 55.1 8e-08
At4g21180_2 49.7 3e-06
At3g28870_2 48.5 6e-06
CE24546_2 45.8 4e-05
SPBC13G1.10c 39.3 0.005
SPBC16H5.08c 29.6 3.6
CE28231 28.9 6.3
Hs22052035 28.5 6.8
Hs16445393 28.5 7.1
> At2g42270
Length=2172
Score = 163 bits (412), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 78/179 (43%), Positives = 117/179 (65%), Gaps = 5/179 (2%)
Query 6 SALRQLPHFSTDLVKAAN---AMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNR 62
S L QLPHF+ DL K + ++ +FD + MED +R++LL ++ AQL ++A+ NR
Sbjct 1986 SMLLQLPHFTKDLAKRCHENPGNNIETIFDLVEMEDDKRQELL-QMSDAQLLDIARFCNR 2044
Query 63 YPVISLEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQ 122
+P I L +E+ + +SPG++I V LERD+ VGPV AP +PK KEE WWLV+G+
Sbjct 2045 FPNIDLTYEIVGSNEVSPGKDITLQVLLERDMEGRTEVGPVDAPRYPKTKEEGWWLVVGE 2104
Query 123 SASNGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRVR 181
+ +N L AIKRIS+ +++ +KL F P G+ + L+ MCDSY+G DQE+ F + V+
Sbjct 2105 AKTNQLMAIKRISLQRKAQ-VKLEFAVPTETGEKSYTLYFMCDSYLGCDQEYSFTVDVK 2162
> At1g20960
Length=2171
Score = 160 bits (404), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 77/179 (43%), Positives = 115/179 (64%), Gaps = 5/179 (2%)
Query 6 SALRQLPHFSTDLVKAAN---AMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNR 62
S L QLPHF+ DL K ++ VFD + MED +R++LL ++ AQL ++A+ NR
Sbjct 1985 SMLLQLPHFTKDLAKRCQENPGKNIETVFDLVEMEDEERQELL-KMSDAQLLDIARFCNR 2043
Query 63 YPVISLEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQ 122
+P I L +E+ +E ++PG+ + V LERD+ VGPV + +PK KEE WWLV+G
Sbjct 2044 FPNIDLTYEIVGSEEVNPGKEVTLQVMLERDMEGRTEVGPVDSLRYPKTKEEGWWLVVGD 2103
Query 123 SASNGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRVR 181
+ +N L AIKR+S+ ++ +KL F AP PG+ + L+ MCDSY+G DQE+ F + V+
Sbjct 2104 TKTNQLLAIKRVSLQRKVK-VKLDFTAPSEPGEKSYTLYFMCDSYLGCDQEYSFSVDVK 2161
> Hs22042312
Length=2136
Score = 157 bits (396), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 79/176 (44%), Positives = 116/176 (65%), Gaps = 6/176 (3%)
Query 6 SALRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPV 65
S L+QLPHF+++ +K V+ VFD M MED +R LL LT +Q+ +VA+ NRYP
Sbjct 1958 SYLKQLPHFTSEHIKRCTDKGVESVFDIMEMEDEERNALL-QLTDSQIADVARFCNRYPN 2016
Query 66 ISLEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQSAS 125
I L +E+ ++I G + VQLER+ + GPV APLFP+++EE WW+VIG + S
Sbjct 2017 IELSYEVVDKDSIRSGGPVVVLVQLERE---EEVTGPVIAPLFPQKREEGWWVVIGDAKS 2073
Query 126 NGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRVR 181
N L +IKR+++ +Q + +KL F AP T G H + L+ M D+Y+G DQE+KF + V+
Sbjct 2074 NSLISIKRLTL-QQKAKVKLDFVAPAT-GAHNYTLYFMSDAYMGCDQEYKFSVDVK 2127
> CE21971
Length=2145
Score = 135 bits (340), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 77/174 (44%), Positives = 106/174 (60%), Gaps = 5/174 (2%)
Query 8 LRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPVIS 67
L+QLPH S L++ A A EV VF+ + +E+ R +L + A+L +VA+ N YP I
Sbjct 1959 LKQLPHCSAALLERAKAKEVTSVFELLELENDDRSDIL-QMEGAELADVARFCNHYPSIE 2017
Query 68 LEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFP-KEKEEQWWLVIGQSASN 126
+ EL + + ++ +N+ V LERD PV APLFP K KEE WWLVIG S SN
Sbjct 2018 VATEL-ENDVVTSNDNLMLAVSLERDNDIDGLAPPVVAPLFPQKRKEEGWWLVIGDSESN 2076
Query 127 GLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRV 180
L IKR+ + ++SS ++L F AP PG H+F LF + DSY+GADQE +V
Sbjct 2077 ALLTIKRLVINEKSS-VQLDFAAPR-PGHHKFKLFFISDSYLGADQEFDVAFKV 2128
> YER172c
Length=2163
Score = 128 bits (321), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 63/174 (36%), Positives = 110/174 (63%), Gaps = 4/174 (2%)
Query 8 LRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPVIS 67
LRQ+PHF+ +++ + V+ V+D M +ED +R+++L +LT +QL +VA N YP +
Sbjct 1994 LRQIPHFNNKILEKCKEINVETVYDIMALEDEERDEIL-TLTDSQLAQVAAFVNNYPNVE 2052
Query 68 LEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQSASNG 127
L + L+ ++++ G + T+QL RD+ + V + +P +K E WWLV+G+ +
Sbjct 2053 LTYSLNNSDSLISGVKQKITIQLTRDVEPENL--QVTSEKYPFDKLESWWLVLGEVSKKE 2110
Query 128 LNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRVR 181
L AIK++++ K++ +L F+ P T GKH ++ +CDSY+ AD+E FEI V+
Sbjct 2111 LYAIKKVTLNKETQQYELEFDTP-TSGKHNLTIWCVCDSYLDADKELSFEINVK 2163
> SPAC9.03c
Length=2176
Score = 108 bits (271), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 66/176 (37%), Positives = 100/176 (56%), Gaps = 6/176 (3%)
Query 6 SALRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPV 65
S L+Q+P+F L++ N V DVFD ++++D +R +LL + A L + A+ N+YP
Sbjct 1994 SPLKQIPYFDDALIERCNKEGVHDVFDIIDLDDEKRTELL-HMDNAHLAKCAEFINKYPD 2052
Query 66 ISLEFELSKTENISPGENIQCTVQLERDLADGDTVG-PVYAPLFPKEKEEQWWLVIGQSA 124
I ++FE+ +E++ VQL R+L + + V V AP FP +K E WWLVI S
Sbjct 2053 IDIDFEIEDSEDVHANSPSVLIVQLTRELEEDEEVDTTVIAPYFPAQKTEHWWLVI--SD 2110
Query 125 SNGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRV 180
L AIK+I++ + S T K+ F P G ++ L DSY+G D E +FE V
Sbjct 2111 DKTLLAIKKITLGR-SLTTKMEF-VPPAMGTLKYKLSCFSDSYMGVDYEKEFECNV 2164
> CE20305
Length=1798
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 44/148 (29%), Positives = 69/148 (46%), Gaps = 16/148 (10%)
Query 48 LTQAQLREVAKASNRYPVISLEF--------------ELSKTENISPGENIQCTVQLERD 93
L +AQ REV KA +P+I+++ E K ++ GE + + +ER
Sbjct 1651 LDEAQSREVLKALCNWPIINMKIMQLVDSRGNCVDIDETKKPVKVTAGEVYKLRIVMER- 1709
Query 94 LADGDTVGPVYAPLFPKEKEEQWWLVIGQ-SASNGLNAIKRISVTKQSSTIKLAFEAPET 152
+ G ++ P +PK K+ W +V+G SA LN ST KL AP T
Sbjct 1710 VGPGKNNSSMHLPQWPKPKQAGWIIVVGNVSADMILNTTTVTGSHSTRSTAKLDIRAPAT 1769
Query 153 PGKHQFVLFLMCDSYIGADQEHKFEIRV 180
G H+ + ++ D Y+G DQE+ + V
Sbjct 1770 KGNHELAVLILSDCYLGIDQEYTLRLDV 1797
> Hs14747366
Length=1866
Score = 69.3 bits (168), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 27/76 (35%), Positives = 44/76 (57%), Gaps = 0/76 (0%)
Query 105 APLFPKEKEEQWWLVIGQSASNGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMC 164
P FPK K+E W+L++G+ L A+KR+ + L+F PE PG++ + L+ M
Sbjct 1768 TPRFPKSKDEGWFLILGEVDKRELIALKRVGYIRNHHVASLSFYTPEIPGRYIYTLYFMS 1827
Query 165 DSYIGADQEHKFEIRV 180
D Y+G DQ++ + V
Sbjct 1828 DCYLGLDQQYDIYLNV 1843
> 7300033
Length=1809
Score = 66.6 bits (161), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 60/109 (55%), Gaps = 7/109 (6%)
Query 77 NISPGENIQCTVQLERDLADGDTVG-----PVYAPLFPKEKEEQWWLVIGQSASNGLNAI 131
++ E+ V L+R G G V+ P +PK K E W+L +G A++ L A+
Sbjct 1695 SLHANEDYVLIVNLQRLNVSGQRRGGGQSYTVHCPKYPKPKNEAWFLTLGSQANDELLAM 1754
Query 132 KRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEH--KFEI 178
KR+S+ Q T +++F+A G+ Q L+LM D +G DQ++ +FEI
Sbjct 1755 KRVSIRGQRCTNRISFQATPRLGRLQLTLYLMSDCLMGFDQQYDLQFEI 1803
> Hs6005872_2
Length=518
Score = 65.9 bits (159), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 45/80 (56%), Gaps = 1/80 (1%)
Query 102 PVYAPLFPKEKEEQWWLVIGQSASNGLNAIK-RISVTKQSSTIKLAFEAPETPGKHQFVL 160
PVY+ FP+EK+E WWL I L ++ + K + ++L F AP PG +Q+ +
Sbjct 395 PVYSLYFPEEKQEWWWLYIADRKEQTLISMPYHVCTLKDTEEVELKFPAPGKPGNYQYTV 454
Query 161 FLMCDSYIGADQEHKFEIRV 180
FL DSY+G DQ ++ V
Sbjct 455 FLRSDSYMGLDQIKPLKLEV 474
> At1g79940_2
Length=480
Score = 62.8 bits (151), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 54/221 (24%), Positives = 88/221 (39%), Gaps = 49/221 (22%)
Query 6 SALRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLL---ASLTQAQLREVAKASNR 62
S QLPHFS +VK +VK D M R +LL A L+ + ++ K
Sbjct 182 SPFLQLPHFSDAVVKKIARKKVKSFQDLQEMRLEDRSELLTQVAGLSATDVEDIEKVLEM 241
Query 63 YPVISLEF--ELSKTENISPGE--NIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWL 118
P I+++ E E I G+ +Q V L+R +G +AP FP KEE +W+
Sbjct 242 MPSITVDITCETEGEEGIQEGDIVTLQAWVTLKR--PNGLVGALPHAPYFPFHKEENYWV 299
Query 119 VIGQSASNGLNAIKRISVTKQSSTIKLA-------------------------------- 146
++ S SN + +++S + I A
Sbjct 300 LLADSVSNNVWFSQKVSFLDEGGAITAASKAISESMEGSGAGVKETNDAVREAIEKVKGG 359
Query 147 -------FEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRV 180
+AP G + F +CD++IG D++ +++V
Sbjct 360 SRLVMGKLQAP-AEGTYNLTCFCLCDTWIGCDKKQALKVKV 399
> 7295249_2
Length=521
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 44/80 (55%), Gaps = 1/80 (1%)
Query 103 VYAPLFPKEKEEQWWLVIGQSASNGL-NAIKRISVTKQSSTIKLAFEAPETPGKHQFVLF 161
V+ P FP+EK+E WW I S L A ++ + I+L F AP PG + F +
Sbjct 400 VHCPYFPEEKQEYWWTYICDRKSRTLLTAPYHVTNLVEKEEIQLKFTAPRWPGVYTFTVC 459
Query 162 LMCDSYIGADQEHKFEIRVR 181
L DSY+G DQ+ + ++ V+
Sbjct 460 LRSDSYLGMDQQQELKLDVQ 479
> SPBC36B7.03
Length=611
Score = 55.1 bits (131), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 46/205 (22%), Positives = 87/205 (42%), Gaps = 42/205 (20%)
Query 8 LRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPVIS 67
L QLPH + VK + + + F+++ + Q + L + ++ QL+E+ + +N P IS
Sbjct 338 LLQLPHLLMEDVKNLSIRNISSIPQFLSLSEEQTKDYLPNYSKNQLKEMREIANGIPRIS 397
Query 68 L---EFELSKTENISPGE------NIQCTVQLE--------------------------- 91
+ + + E I+ G +++C+ E
Sbjct 398 VVAAKVLVDDDEYITVGAIANLILDLKCSYGTEVVPEVSTDGTETATKKDEEDAEKYHQS 457
Query 92 RDLADGD--TVGPVYAPLFPKEKEEQWWLVIGQSASNGLNAIKRISVT---KQSSTIKLA 146
+D+ GD T+ +AP F + + WW+ + N + + S+T K T ++
Sbjct 458 KDVVLGDVETLPYAWAPYFTQHHKTAWWIYVVDPRQNRV-IVPPFSITDIPKTLRTFRIP 516
Query 147 FEAPETPGKHQFVLFLMCDSYIGAD 171
F+ P G F L +M +SY+G D
Sbjct 517 FQVPPVAGTFSFQLHIMSNSYVGED 541
> At5g61140
Length=2137
Score = 55.1 bits (131), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 36/134 (26%), Positives = 65/134 (48%), Gaps = 5/134 (3%)
Query 4 QASALRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRY 63
Q S+L +P + L+ + A + + +N+ RE L + +++ R+
Sbjct 1974 QDSSLWMIPCMNDLLLGSLTARGIHTLHQLLNLP---RETLQSVTENFPASRLSQDLQRF 2030
Query 64 PVISLEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQS 123
P I + L K + S G+ T+++ + AP FPK K+E WWLV+G +
Sbjct 2031 PRIQMNVRLQKKD--SDGKKKPSTLEIRLEKTSKRNSSRALAPRFPKVKDEAWWLVLGDT 2088
Query 124 ASNGLNAIKRISVT 137
+++ L A+KR+S T
Sbjct 2089 STSELFAVKRVSFT 2102
> At4g21180_2
Length=439
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 53/216 (24%), Positives = 94/216 (43%), Gaps = 50/216 (23%)
Query 10 QLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLL---ASLTQAQLREVAKASNRYPV- 65
QLPHF+ + K+ A++VK F + +R KLL SL++ ++++ K P
Sbjct 168 QLPHFNESIAKSI-ALQVKSFQKFQELSLAERSKLLREVVSLSETDVQDIEKVLEMIPSL 226
Query 66 -ISLEFELSKTENISPGE--NIQCTVQLERDLADGDTVGPV-YAPLFPKEKEEQWWLVIG 121
I++ + E I G+ +Q + L+R +G + ++P FP KEE +W+++
Sbjct 227 KINVTCKTEGEEGIQEGDIMTVQAWITLKRP---NGLIGAIPHSPYFPFHKEENFWVLLA 283
Query 122 QS--------------------ASNGL---------------NAIKRISVTKQSSTIKLA 146
S ASN + +A+K +V K S +L
Sbjct 284 DSNHVWFFQKVKFMDEAGAIAAASNTITETMEPLGASVKETNDAVKE-AVEKVKSGSRLV 342
Query 147 FEAPETPGKHQFVL--FLMCDSYIGADQEHKFEIRV 180
PG+ + L F + D++IG DQ+ ++ V
Sbjct 343 MGRLLAPGEGTYNLTCFCLSDTWIGCDQKTSLKVEV 378
> At3g28870_2
Length=59
Score = 48.5 bits (114), Expect = 6e-06, Method: Composition-based stats.
Identities = 17/38 (44%), Positives = 25/38 (65%), Gaps = 0/38 (0%)
Query 141 STIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEI 178
+ +KL F P PG+ + L+ MCDSY+G DQE+ F +
Sbjct 9 AKVKLDFTVPSEPGEKSYTLYFMCDSYLGCDQEYSFSV 46
> CE24546_2
Length=535
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 40/84 (47%), Gaps = 5/84 (5%)
Query 103 VYAPLFPKEKEEQWWLVIG-----QSASNGLNAIKRISVTKQSSTIKLAFEAPETPGKHQ 157
V+AP FP EK E WW+ + + + L + + TI + F AP G +
Sbjct 415 VHAPFFPTEKFEWWWITVAYVDKKEKSRQLLTFPQLVKTLIDEQTIDIRFAAPPHKGIYT 474
Query 158 FVLFLMCDSYIGADQEHKFEIRVR 181
+ L + DSY+ A+ F+I V+
Sbjct 475 YNLSVKSDSYMDAEYSVDFKIDVK 498
> SPBC13G1.10c
Length=1935
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 39/71 (54%), Gaps = 6/71 (8%)
Query 103 VYAPLFPKEKEEQWWLVIGQSASNGLNAIKRISVTKQSS------TIKLAFEAPETPGKH 156
++APLFPK + E ++++I S + L AI+R S + + +++++ + P T
Sbjct 1853 IFAPLFPKPQSEGFFVLIIDSETQELFAIRRASFAGRRNDDSIRLSLRISMDIPPTCRNR 1912
Query 157 QFVLFLMCDSY 167
+ ++CD Y
Sbjct 1913 NVKVMVVCDGY 1923
> SPBC16H5.08c
Length=618
Score = 29.6 bits (65), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 22/97 (22%), Positives = 42/97 (43%), Gaps = 1/97 (1%)
Query 65 VISLEFELSKTENISPGENIQCTV-QLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQS 123
+I +E +S+ + + Q + QL D + + + Y P FP+ + +QW V+G+
Sbjct 440 LIPIEGNVSRYSGLKMAKYSQHSADQLPYDKSPLEYIMDTYKPKFPERELQQWRSVLGKF 499
Query 124 ASNGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVL 160
+GL+ I ++ F A H +L
Sbjct 500 GLSGLHQTSEIRTLSDGLKSRVVFAALALEQPHILLL 536
> CE28231
Length=762
Score = 28.9 bits (63), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 20/76 (26%), Positives = 38/76 (50%), Gaps = 1/76 (1%)
Query 20 KAANAMEVK-DVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPVISLEFELSKTENI 78
+AA + V+ ++FD M ++ KL+ T QL + + + + V L+ + SK
Sbjct 374 RAAKDLTVELEIFDKMFLQRKDLIKLIGKATLDQLLPPSNSKSAHNVAELQLKASKDSEF 433
Query 79 SPGENIQCTVQLERDL 94
+ N+ +V L R+L
Sbjct 434 TAKVNVTTSVNLNREL 449
> Hs22052035
Length=431
Score = 28.5 bits (62), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 34/70 (48%), Gaps = 0/70 (0%)
Query 24 AMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPVISLEFELSKTENISPGEN 83
A E+ D + ++G ++LAS T+ QLRE+ KA SLE ++ +
Sbjct 203 AKELHDAMKGLGTKEGVIIEILASRTKNQLREIMKAYEEDYGSSLEEDIQADTSGYLERI 262
Query 84 IQCTVQLERD 93
+ C +Q RD
Sbjct 263 LVCLLQGSRD 272
> Hs16445393
Length=794
Score = 28.5 bits (62), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 64 PVISLEFELSKTENISPGENIQCTVQLERDLA 95
P IS+ +E + EN PG+ IQ +RDL+
Sbjct 487 PEISVPYETAVCENAKPGQIIQIVSAADRDLS 518
Lambda K H
0.316 0.131 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2842629034
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40