bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
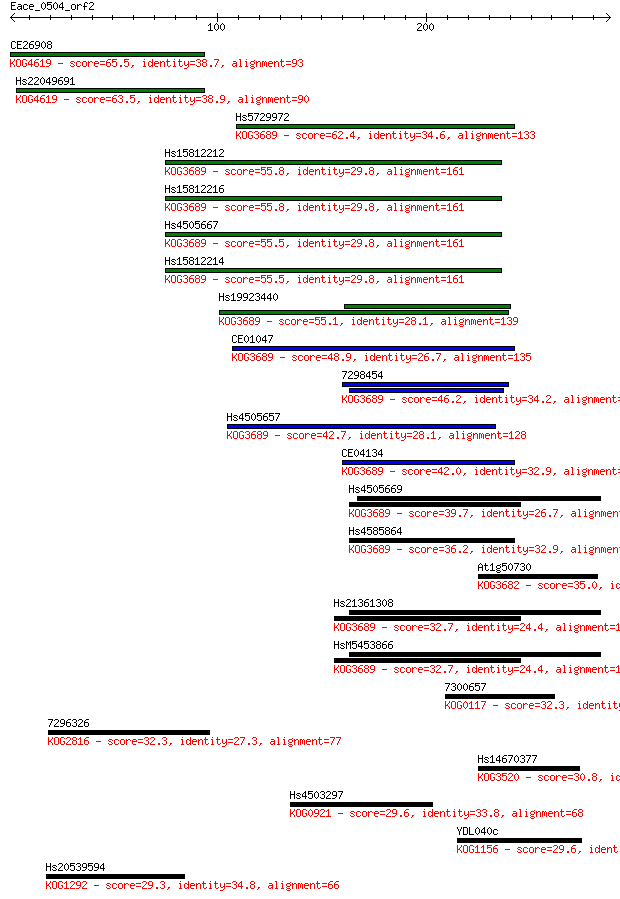
Query= Eace_0504_orf2
Length=287
Score E
Sequences producing significant alignments: (Bits) Value
CE26908 65.5 1e-10
Hs22049691 63.5 4e-10
Hs5729972 62.4 1e-09
Hs15812212 55.8 1e-07
Hs15812216 55.8 1e-07
Hs4505667 55.5 1e-07
Hs15812214 55.5 1e-07
Hs19923440 55.1 2e-07
CE01047 48.9 1e-05
7298454 46.2 8e-05
Hs4505657 42.7 8e-04
CE04134 42.0 0.001
Hs4505669 39.7 0.008
Hs4585864 36.2 0.072
At1g50730 35.0 0.20
Hs21361308 32.7 0.85
HsM5453866 32.7 0.87
7300657 32.3 1.1
7296326 32.3 1.3
Hs14670377 30.8 3.6
Hs4503297 29.6 6.9
YDL040c 29.6 8.4
Hs20539594 29.3 9.6
> CE26908
Length=224
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 36/93 (38%), Positives = 49/93 (52%), Gaps = 0/93 (0%)
Query 1 QLIMIRLGDLLDTTLGVKFGLATLSAAAVGQLCSDTAGVLFGSTIESCASRLGFPPPNLT 60
+IMI G+ +D LG ++T++AAA+G L SD AGV +E R+G P LT
Sbjct 110 NMIMILAGEYIDQKLGAVLAISTMAAAALGNLISDIAGVGLAHYVEVAVQRVGIKHPVLT 169
Query 61 AAQRASAAFGIVKTFGAASGVCCGCLLGMLQLL 93
+ Q S A+G+ GCLLGM LL
Sbjct 170 SQQLDSGKARFTTNAARAAGLTVGCLLGMFPLL 202
> Hs22049691
Length=105
Score = 63.5 bits (153), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 35/90 (38%), Positives = 52/90 (57%), Gaps = 0/90 (0%)
Query 4 MIRLGDLLDTTLGVKFGLATLSAAAVGQLCSDTAGVLFGSTIESCASRLGFPPPNLTAAQ 63
MI G ++ ++G+ G++T++AAA+G L SD AG+ +E+ ASRLG P+LT Q
Sbjct 1 MIVAGTHIEMSIGIILGISTMAAAALGNLVSDLAGLGLAGYVEALASRLGLSIPDLTPKQ 60
Query 64 RASAAFGIVKTFGAASGVCCGCLLGMLQLL 93
+ G A GV GC+LGM L+
Sbjct 61 VDMWQTRLSTHLGKAVGVTIGCILGMFPLI 90
> Hs5729972
Length=779
Score = 62.4 bits (150), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 46/145 (31%), Positives = 69/145 (47%), Gaps = 18/145 (12%)
Query 109 LDAIFETVLIEGPKLFHCERAALFVYDPSRDEVWS-----------KAVFGEGSAIRMHK 157
+D++ E ++I L + +R ALF D E++S K VF + IR
Sbjct 267 IDSLLEHIMIYAKNLVNADRCALFQVDHKNKELYSDLFDIGEEKEGKPVFKKTKEIRF-- 324
Query 158 TKNKCITTWVMQNKTILNCPEASEDPRFNPEFDENFEQKTRSVLAAPVMDRSGEVIAALV 217
+ K I V + +LN P+A DPRFN E D TR++L P++ R G VI +
Sbjct 325 SIEKGIAGQVARTGEVLNIPDAYADPRFNREVDLYTGYTTRNILCMPIVSR-GSVIGVVQ 383
Query 218 FFNKHPQFTGR-FSEDDERMVEMMS 241
NK +G FS+ DE +M +
Sbjct 384 MVNK---ISGSAFSKTDENNFKMFA 405
> Hs15812212
Length=833
Score = 55.8 bits (133), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 48/174 (27%), Positives = 72/174 (41%), Gaps = 26/174 (14%)
Query 75 FGAASGVCCGCLLGMLQLLVIDLDKADRLKHQQQLDAIFETVLIEGPKLFHCERAALFVY 134
F G C LL +++ + LD + A+ + + L +R +LF+
Sbjct 99 FDHDEGDQCSRLLELVKDISSHLD----------VTALCHKIFLHIHGLISADRYSLFLV 148
Query 135 --DPSRDEVWSKAVFGEGSAIRMHKTKNKCIT-TWVMQNKTI----------LNCPEASE 181
D S D+ +F + + N CI W NK I LN +A E
Sbjct 149 CEDSSNDKFLISRLFDVAEGSTLEEVSNNCIRLEW---NKGIVGHVAALGEPLNIKDAYE 205
Query 182 DPRFNPEFDENFEQKTRSVLAAPVMDRSGEVIAALVFFNKHPQFTGRFSEDDER 235
DPRFN E D+ KT+S+L P+ + EV+ NK G F+E DE+
Sbjct 206 DPRFNAEVDQITGYKTQSILCMPIKNHREEVVGVAQAINKKSGNGGTFTEKDEK 259
> Hs15812216
Length=823
Score = 55.8 bits (133), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 48/174 (27%), Positives = 72/174 (41%), Gaps = 26/174 (14%)
Query 75 FGAASGVCCGCLLGMLQLLVIDLDKADRLKHQQQLDAIFETVLIEGPKLFHCERAALFVY 134
F G C LL +++ + LD + A+ + + L +R +LF+
Sbjct 89 FDHDEGDQCSRLLELVKDISSHLD----------VTALCHKIFLHIHGLISADRYSLFLV 138
Query 135 --DPSRDEVWSKAVFGEGSAIRMHKTKNKCIT-TWVMQNKTI----------LNCPEASE 181
D S D+ +F + + N CI W NK I LN +A E
Sbjct 139 CEDSSNDKFLISRLFDVAEGSTLEEVSNNCIRLEW---NKGIVGHVAALGEPLNIKDAYE 195
Query 182 DPRFNPEFDENFEQKTRSVLAAPVMDRSGEVIAALVFFNKHPQFTGRFSEDDER 235
DPRFN E D+ KT+S+L P+ + EV+ NK G F+E DE+
Sbjct 196 DPRFNAEVDQITGYKTQSILCMPIKNHREEVVGVAQAINKKSGNGGTFTEKDEK 249
> Hs4505667
Length=875
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 48/174 (27%), Positives = 72/174 (41%), Gaps = 26/174 (14%)
Query 75 FGAASGVCCGCLLGMLQLLVIDLDKADRLKHQQQLDAIFETVLIEGPKLFHCERAALFVY 134
F G C LL +++ + LD + A+ + + L +R +LF+
Sbjct 141 FDHDEGDQCSRLLELVKDISSHLD----------VTALCHKIFLHIHGLISADRYSLFLV 190
Query 135 --DPSRDEVWSKAVFGEGSAIRMHKTKNKCIT-TWVMQNKTI----------LNCPEASE 181
D S D+ +F + + N CI W NK I LN +A E
Sbjct 191 CEDSSNDKFLISRLFDVAEGSTLEEVSNNCIRLEW---NKGIVGHVAALGEPLNIKDAYE 247
Query 182 DPRFNPEFDENFEQKTRSVLAAPVMDRSGEVIAALVFFNKHPQFTGRFSEDDER 235
DPRFN E D+ KT+S+L P+ + EV+ NK G F+E DE+
Sbjct 248 DPRFNAEVDQITGYKTQSILCMPIKNHREEVVGVAQAINKKSGNGGTFTEKDEK 301
> Hs15812214
Length=865
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 48/174 (27%), Positives = 72/174 (41%), Gaps = 26/174 (14%)
Query 75 FGAASGVCCGCLLGMLQLLVIDLDKADRLKHQQQLDAIFETVLIEGPKLFHCERAALFVY 134
F G C LL +++ + LD + A+ + + L +R +LF+
Sbjct 141 FDHDEGDQCSRLLELVKDISSHLD----------VTALCHKIFLHIHGLISADRYSLFLV 190
Query 135 --DPSRDEVWSKAVFGEGSAIRMHKTKNKCIT-TWVMQNKTI----------LNCPEASE 181
D S D+ +F + + N CI W NK I LN +A E
Sbjct 191 CEDSSNDKFLISRLFDVAEGSTLEEVSNNCIRLEW---NKGIVGHVAALGEPLNIKDAYE 247
Query 182 DPRFNPEFDENFEQKTRSVLAAPVMDRSGEVIAALVFFNKHPQFTGRFSEDDER 235
DPRFN E D+ KT+S+L P+ + EV+ NK G F+E DE+
Sbjct 248 DPRFNAEVDQITGYKTQSILCMPIKNHREEVVGVAQAINKKSGNGGTFTEKDEK 301
> Hs19923440
Length=934
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 46/79 (58%), Gaps = 1/79 (1%)
Query 161 KCITTWVMQNKTILNCPEASEDPRFNPEFDENFEQKTRSVLAAPVMDRSGEVIAALVFFN 220
K I +V ++ +N P+A +D RFN E D+ KT+S+L P+ GE+I N
Sbjct 284 KGIIGYVGEHGETVNIPDAYQDRRFNDEIDKLTGYKTKSLLCMPIRSSDGEIIGVAQAIN 343
Query 221 KHPQFTGRFSEDDERMVEM 239
K P+ F+EDDE++++M
Sbjct 344 KIPE-GAPFTEDDEKVMQM 361
Score = 40.0 bits (92), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 32/157 (20%), Positives = 62/157 (39%), Gaps = 22/157 (14%)
Query 101 DRLKHQQQLDAIFETVLIEGPKLFHCERAALFVYDPSRD---------EVWSKAVFGEGS 151
D + Q L+ I + ++ L CER ++ + + E+ S +
Sbjct 395 DLFEEQTDLEKIVKKIMHRAQTLLKCERCSVLLLEDIESPVVKFTKSFELMSPKCSADAE 454
Query 152 AIRMHKTKNKCITTWVMQNKTI---------LNCPEASEDPRFNPEFDENFEQKTRSVLA 202
+ + W++ N +N +A +DPRF+ E D+ RSVL
Sbjct 455 NSFKESMEKSSYSDWLINNSIAELVASTGLPVNISDAYQDPRFDAEADQISGFHIRSVLC 514
Query 203 APVMDRSGEVIAALVFFNKHPQFTGR-FSEDDERMVE 238
P+ + + ++I N+ G+ F + D+R+ E
Sbjct 515 VPIWNSNHQIIGVAQVLNR---LDGKPFDDADQRLFE 548
> CE01047
Length=918
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 36/137 (26%), Positives = 61/137 (44%), Gaps = 9/137 (6%)
Query 107 QQLDAIFETVLIEGPKLFHCERAALFVYDPSRDEVWSKAVFGEGSAIRMHKTKN--KCIT 164
L A+ ++ E K E A+F++D ++ +F + + K + I
Sbjct 235 NNLSALISCIIAEAKKNTEAEDYAVFLHDEDNKQM---VLFNNETMLMTGKKFDMGYGIV 291
Query 165 TWVMQNKTILNCPEASEDPRFNPEFDENFEQKTRSVLAAPVMDRSGEVIAALVFFNKHPQ 224
V +N + S P FN E DE F K R+++A P++D S +I +V +NK
Sbjct 292 GKVASTMRTMNIRDVSRCPFFNEEIDEQFSIKARNLIAFPLIDSSCSLIGVIVLYNKE-- 349
Query 225 FTGRFSEDDERMVEMMS 241
FS DE+ ++ S
Sbjct 350 --NGFSRHDEKYIKRFS 364
> 7298454
Length=1284
Score = 46.2 bits (108), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 39/79 (49%), Gaps = 2/79 (2%)
Query 160 NKCITTWVMQNKTILNCPEASEDPRFNPEFDENFEQKTRSVLAAPVMDRSGEVIAALVFF 219
N IT V +N P A ED RF+ DEN K RS+L + + G++I +
Sbjct 475 NIGITGHVATTGETVNVPNAYEDDRFDASVDENSCFKHRSILCMAIKNSLGQIIGVIQLI 534
Query 220 NKHPQFTGRFSEDDERMVE 238
NK + F+++DE VE
Sbjct 535 NKFNELD--FTKNDENFVE 551
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 40/75 (53%), Gaps = 4/75 (5%)
Query 163 ITTWVMQNKTILNCPEASEDPRFNPEFDENFEQKTRSVLAAPVMDRSGEVIAALVFFNKH 222
I V ++ +N P+A +D RFN E D +T+++L P+ D SG+VI NK
Sbjct 296 IAGHVAESGEPVNIPDAYQDERFNCEIDSLTGYRTKALLCMPIKDSSGDVIGVAQVINK- 354
Query 223 PQFTGR-FSEDDERM 236
G FSE DE++
Sbjct 355 --MNGECFSEIDEKV 367
> Hs4505657
Length=941
Score = 42.7 bits (99), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 36/135 (26%), Positives = 63/135 (46%), Gaps = 12/135 (8%)
Query 105 HQQQLDAIFETVLIEGPKLFHCERAALFVYDPSRDEVWSKAVFG-----EGSAIRMHKTK 159
H + + + ++ E L + E ++F+ D ++E+ +K G E IR+
Sbjct 406 HLDDVSVLLQEIITEARNLSNAEICSVFLLD--QNELVAKVFDGGVVDDESYEIRI--PA 461
Query 160 NKCITTWVMQNKTILNCPEASEDPRFNPEFDENFEQKTRSVLAAPVMDRSGEVIAALVFF 219
++ I V ILN P+A P F D++ +TR++L P+ + + EVI
Sbjct 462 DQGIAGHVATTGQILNIPDAYAHPLFYRGVDDSTGFRTRNILCFPIKNENQEVIGVAELV 521
Query 220 NK--HPQFTGRFSED 232
NK P F+ +F ED
Sbjct 522 NKINGPWFS-KFDED 535
> CE04134
Length=393
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 39/82 (47%), Gaps = 4/82 (4%)
Query 160 NKCITTWVMQNKTILNCPEASEDPRFNPEFDENFEQKTRSVLAAPVMDRSGEVIAALVFF 219
+K I +V LN A ED RFN + D T+++L P++ R G VI +
Sbjct 2 SKGIAGYVASTGEGLNIENAYEDERFNADVDSKTGYTTKTILCMPILIR-GIVIGVVQMV 60
Query 220 NKHPQFTGRFSEDDERMVEMMS 241
NKH G F+ DE E+ +
Sbjct 61 NKHD---GVFTRQDEDAFEIFA 79
> Hs4505669
Length=854
Score = 39.7 bits (91), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 32/122 (26%), Positives = 64/122 (52%), Gaps = 10/122 (8%)
Query 167 VMQNKTILNCPEASEDPRFNPEFDENFEQKTRSVLAAPVMDRSGEVIAALVFFNK--HPQ 224
V Q K ++N + +E P F+ DE + KT+++LA P+M+ +V+A ++ NK P
Sbjct 142 VAQTKKMVNVEDVAECPHFSSFADELTDYKTKNMLATPIMN-GKDVVAVIMAVNKLNGPF 200
Query 225 FTGRFSEDDE---RMVEMMSRHMQIFMEMFEYGAAEDRKMIYLPEPKEAF-KLSGVETEK 280
FT SED++ + + + +++I+ + + R + L + F +L+ +E +
Sbjct 201 FT---SEDEDVFLKYLNFATLYLKIYHLSYLHNCETRRGQVLLWSANKVFEELTDIERQF 257
Query 281 EK 282
K
Sbjct 258 HK 259
Score = 29.6 bits (65), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 21/87 (24%), Positives = 46/87 (52%), Gaps = 10/87 (11%)
Query 163 ITTWVMQNKTILNCPEASEDPRFNPEFDENFEQKT----RSVLAAPVMDRSGEVIAALVF 218
+ ++V ++ I N AS D F +F E + ++VL+ P++++ E++ F
Sbjct 344 LPSYVAESGFICNIMNASADEMF--KFQEGALDDSGWLIKNVLSMPIVNKKEEIVGVATF 401
Query 219 FNKHPQFTGR-FSEDDERMVEMMSRHM 244
+N+ G+ F E DE ++E +++ +
Sbjct 402 YNRK---DGKPFDEQDEVLMESLTQFL 425
> Hs4585864
Length=860
Score = 36.2 bits (82), Expect = 0.072, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 43/81 (53%), Gaps = 6/81 (7%)
Query 163 ITTWVMQNKTILNCPEASEDPRFNPEFDENFEQKTRSVLAAPVMDRSGEVIAALVFFNK- 221
I V +K I N P ED F D E KT+++LA+P+M+ +V+A ++ NK
Sbjct 140 IVGHVAHSKKIANVPNTEEDEHFCDFVDILTEYKTKNILASPIMN-GKDVVAIIMAVNKV 198
Query 222 -HPQFTGRFSEDDERMVEMMS 241
FT R D+E +++ ++
Sbjct 199 DGSHFTKR---DEEILLKYLN 216
> At1g50730
Length=1013
Score = 35.0 bits (79), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 33/57 (57%), Gaps = 6/57 (10%)
Query 225 FTGRFSEDDERMVEMMSRHMQIFMEMFEYGAAEDRKMIYLPEPKEAFKLSGVETEKE 281
F +D+ + ++SR F+EM +YGA +R +++L E +EAF +G+ KE
Sbjct 612 FVNIKDDDNRQTSHLISR----FVEMVDYGAEMERHLLFLAECREAF--NGIHELKE 662
> Hs21361308
Length=858
Score = 32.7 bits (73), Expect = 0.85, Method: Compositional matrix adjust.
Identities = 29/125 (23%), Positives = 53/125 (42%), Gaps = 8/125 (6%)
Query 163 ITTWVMQNKTILNCPEASEDPRFNPEFDENFEQKTRSVLAAPVMDRSGEVIAALVFFNKH 222
I W K N P+ ++ F+ D+ T+++LA P++ EV+A ++ NK
Sbjct 142 IVGWAAHTKKTHNVPDVKKNSHFSDFMDKQTGYVTKNLLATPIV-VGKEVLAVIMAVNKV 200
Query 223 PQFTGRFSEDDERMVEMMSRHMQIFMEM----FEYGAAEDRKMIYLPEPKEAF-KLSGVE 277
FS+ DE + + I + + + Y R I + + F +L+ VE
Sbjct 201 N--ASEFSKQDEEVFSKYLNFVSIILRLHHTSYMYNIESRRSQILMWSANKVFEELTDVE 258
Query 278 TEKEK 282
+ K
Sbjct 259 RQFHK 263
Score = 29.3 bits (64), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 23/94 (24%), Positives = 46/94 (48%), Gaps = 10/94 (10%)
Query 156 HKTKNKCITTWVMQNKTILNCPEASEDPRFNPEFDENFEQKT----RSVLAAPVMDRSGE 211
H T + T+V +N I N A D F F + +T ++VL+ P++++ +
Sbjct 341 HWTLISGLPTYVAENGFICNMMNAPADEYFT--FPKGPVDETGWVIKNVLSLPIVNKKED 398
Query 212 VIAALVFFNKHPQFTGR-FSEDDERMVEMMSRHM 244
++ F+N+ G+ F E DE + E +++ +
Sbjct 399 IVGVATFYNRK---DGKPFDEHDEYITETLTQFL 429
> HsM5453866
Length=858
Score = 32.7 bits (73), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 29/125 (23%), Positives = 53/125 (42%), Gaps = 8/125 (6%)
Query 163 ITTWVMQNKTILNCPEASEDPRFNPEFDENFEQKTRSVLAAPVMDRSGEVIAALVFFNKH 222
I W K N P+ ++ F+ D+ T+++LA P++ EV+A ++ NK
Sbjct 142 IVGWAAHTKKTHNVPDVKKNSHFSDFMDKQTGYVTKNLLATPIV-VGKEVLAVIMAVNKV 200
Query 223 PQFTGRFSEDDERMVEMMSRHMQIFMEM----FEYGAAEDRKMIYLPEPKEAF-KLSGVE 277
FS+ DE + + I + + + Y R I + + F +L+ VE
Sbjct 201 N--ASEFSKQDEEVFSKYLNFVSIILRLHHTSYMYNIESRRSQILMWSANKVFEELTDVE 258
Query 278 TEKEK 282
+ K
Sbjct 259 RQFHK 263
Score = 29.6 bits (65), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 23/94 (24%), Positives = 46/94 (48%), Gaps = 10/94 (10%)
Query 156 HKTKNKCITTWVMQNKTILNCPEASEDPRFNPEFDENFEQKT----RSVLAAPVMDRSGE 211
H T + T+V +N I N A D F F + +T ++VL+ P++++ +
Sbjct 341 HWTLISGLPTYVAENGFICNMMNAPADEYFT--FQKGPVDETGWVIKNVLSLPIVNKKED 398
Query 212 VIAALVFFNKHPQFTGR-FSEDDERMVEMMSRHM 244
++ F+N+ G+ F E DE + E +++ +
Sbjct 399 IVGVATFYNRK---DGKPFDEHDEYITETLTQFL 429
> 7300657
Length=673
Score = 32.3 bits (72), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 15/54 (27%), Positives = 28/54 (51%), Gaps = 2/54 (3%)
Query 209 SGEVIAALVFFNKHPQFTGRFSEDDER--MVEMMSRHMQIFMEMFEYGAAEDRK 260
+G+ I + FN H F G ++ +R ++E S+H E+ Y + +D+K
Sbjct 269 TGKKIGVTISFNNHRLFVGNIPKNRDRDELIEEFSKHAPGLYEVIIYSSPDDKK 322
> 7296326
Length=504
Score = 32.3 bits (72), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 21/79 (26%), Positives = 39/79 (49%), Gaps = 2/79 (2%)
Query 19 FGLATLSAAAVGQLCSDTAGVLFGSTIESCASRLGFPPPNLTAAQRASAAFG--IVKTFG 76
F + T+S+ S A V +T+E+ + GF + A S + G ++K++G
Sbjct 115 FAILTVSSICGSTYSSSLAYVADTTTVENRSKGYGFVAASFGAGIAFSPSLGNYLMKSYG 174
Query 77 AASGVCCGCLLGMLQLLVI 95
+AS + + GM+ +L I
Sbjct 175 SASVILIATITGMINILFI 193
> Hs14670377
Length=1015
Score = 30.8 bits (68), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 25/48 (52%), Gaps = 5/48 (10%)
Query 225 FTGRFSEDDERMVEMMSRHMQIFMEMFEYGAAEDRKMIYLPEPKEAFK 272
F R S D+ + + + QI++EM E G ED LP+P+ F+
Sbjct 470 FQERLSMKDQLIAQSLLEKQQIYLEMAEMGGLED-----LPQPRGLFR 512
> Hs4503297
Length=1279
Score = 29.6 bits (65), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 38/72 (52%), Gaps = 4/72 (5%)
Query 135 DPSRDEVWSKAVFGEGSAIRMHKTKNKCITTW---VMQNKTILNCPEASEDPRF-NPEFD 190
D + D V S FG + HK K K +TT + +K+ +NCP +S+D + +P F
Sbjct 984 DNNLDVVISLLAFGVYPNVCYHKEKRKILTTEGRNALIHKSSVNCPFSSQDMNYPSPFFV 1043
Query 191 ENFEQKTRSVLA 202
+ +TR++ A
Sbjct 1044 FGEKIRTRAISA 1055
> YDL040c
Length=854
Score = 29.6 bits (65), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 31/61 (50%), Gaps = 2/61 (3%)
Query 215 ALVFFNKHPQFTGRFSED--DERMVEMMSRHMQIFMEMFEYGAAEDRKMIYLPEPKEAFK 272
AL FN P+F +F +D D M + ++EM E+G A K +Y+ KEA K
Sbjct 554 ALKRFNAIPKFYKQFEDDQLDFHSYCMRKGTPRAYLEMLEWGKALYTKPMYVRAMKEASK 613
Query 273 L 273
L
Sbjct 614 L 614
> Hs20539594
Length=602
Score = 29.3 bits (64), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 33/68 (48%), Gaps = 12/68 (17%)
Query 18 KFGLATLSAAAV-GQLCSDTAGVLFG-STIESCASRLGFPPPNLTAAQRASAAFGIVKTF 75
++GL T++AAAV G + AG++ +CA G PPP + A R GI
Sbjct 314 QWGLPTVTAAAVLGMFSATLAGIIESIGDYYACARLAGAPPPPVHAINR-----GIFT-- 366
Query 76 GAASGVCC 83
G+CC
Sbjct 367 ---EGICC 371
Lambda K H
0.322 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6375248280
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40