bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
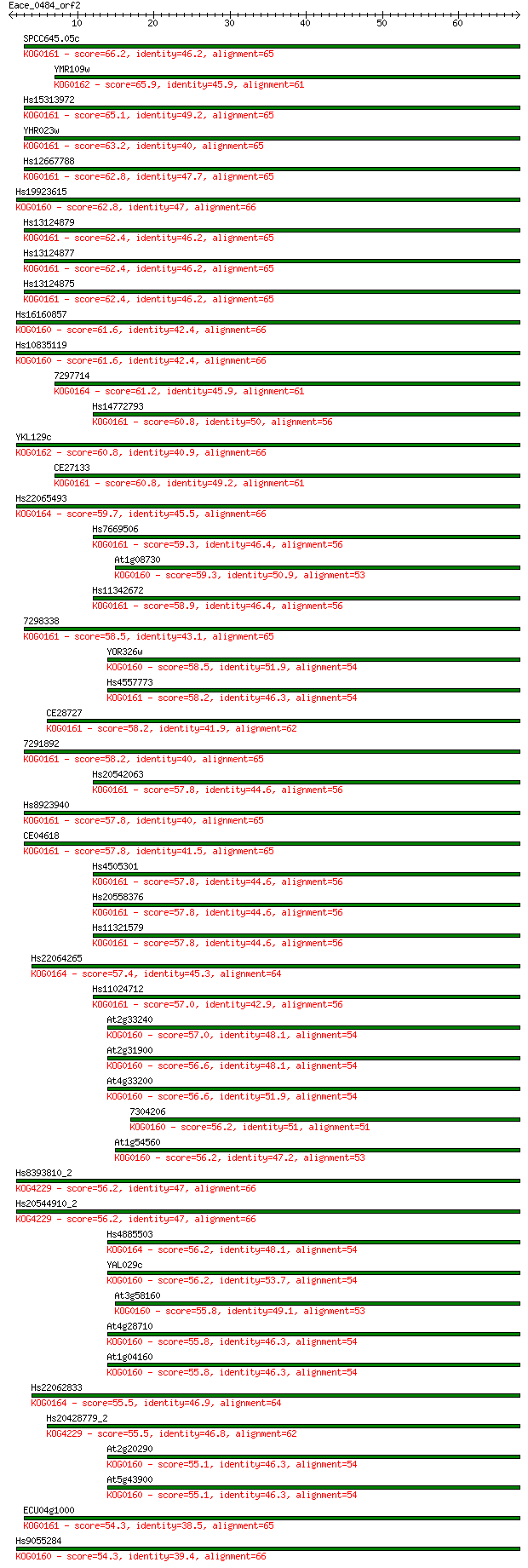
Query= Eace_0484_orf2
Length=67
Score E
Sequences producing significant alignments: (Bits) Value
SPCC645.05c 66.2 1e-11
YMR109w 65.9 2e-11
Hs15313972 65.1 3e-11
YHR023w 63.2 1e-10
Hs12667788 62.8 1e-10
Hs19923615 62.8 2e-10
Hs13124879 62.4 2e-10
Hs13124877 62.4 2e-10
Hs13124875 62.4 2e-10
Hs16160857 61.6 3e-10
Hs10835119 61.6 3e-10
7297714 61.2 4e-10
Hs14772793 60.8 5e-10
YKL129c 60.8 5e-10
CE27133 60.8 6e-10
Hs22065493 59.7 1e-09
Hs7669506 59.3 2e-09
At1g08730 59.3 2e-09
Hs11342672 58.9 2e-09
7298338 58.5 2e-09
YOR326w 58.5 3e-09
Hs4557773 58.2 3e-09
CE28727 58.2 3e-09
7291892 58.2 4e-09
Hs20542063 57.8 4e-09
Hs8923940 57.8 5e-09
CE04618 57.8 5e-09
Hs4505301 57.8 5e-09
Hs20558376 57.8 5e-09
Hs11321579 57.8 5e-09
Hs22064265 57.4 6e-09
Hs11024712 57.0 8e-09
At2g33240 57.0 9e-09
At2g31900 56.6 1e-08
At4g33200 56.6 1e-08
7304206 56.2 1e-08
At1g54560 56.2 1e-08
Hs8393810_2 56.2 1e-08
Hs20544910_2 56.2 1e-08
Hs4885503 56.2 1e-08
YAL029c 56.2 2e-08
At3g58160 55.8 2e-08
At4g28710 55.8 2e-08
At1g04160 55.8 2e-08
Hs22062833 55.5 2e-08
Hs20428779_2 55.5 2e-08
At2g20290 55.1 3e-08
At5g43900 55.1 3e-08
ECU04g1000 54.3 5e-08
Hs9055284 54.3 6e-08
> SPCC645.05c
Length=1526
Score = 66.2 bits (160), Expect = 1e-11, Method: Composition-based stats.
Identities = 30/65 (46%), Positives = 45/65 (69%), Gaps = 0/65 (0%)
Query 3 RTHREADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLE 62
R+ + I+++AK +Y+R FGWLV+ LN ++ + + F+G+LDI GFE+FE NS E
Sbjct 399 RSQTQVISSIEALAKAIYERNFGWLVKRLNTSLNHSNAQSYFIGILDIAGFEIFEKNSFE 458
Query 63 QLLIN 67
QL IN
Sbjct 459 QLCIN 463
> YMR109w
Length=1219
Score = 65.9 bits (159), Expect = 2e-11, Method: Composition-based stats.
Identities = 28/61 (45%), Positives = 43/61 (70%), Gaps = 0/61 (0%)
Query 7 EADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLI 66
+AD + ++AK +Y+ LF W+V +NK++ G +G+LDI+GFE+FEHNS EQ+ I
Sbjct 367 QADAVRDALAKAIYNNLFDWIVSRVNKSLQAFPGAEKSIGILDIYGFEIFEHNSFEQICI 426
Query 67 N 67
N
Sbjct 427 N 427
> Hs15313972
Length=1647
Score = 65.1 bits (157), Expect = 3e-11, Method: Composition-based stats.
Identities = 32/66 (48%), Positives = 44/66 (66%), Gaps = 1/66 (1%)
Query 3 RTHREADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNV-FMGMLDIFGFEVFEHNSL 61
+T +AD ++++AK Y+RLF WLV +NKA+ F+G+LDI GFE+FE NS
Sbjct 83 QTKEQADFAVEALAKATYERLFRWLVHRINKALDRTKRQGASFIGILDIAGFEIFELNSF 142
Query 62 EQLLIN 67
EQL IN
Sbjct 143 EQLCIN 148
> YHR023w
Length=1928
Score = 63.2 bits (152), Expect = 1e-10, Method: Composition-based stats.
Identities = 26/65 (40%), Positives = 47/65 (72%), Gaps = 0/65 (0%)
Query 3 RTHREADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLE 62
+ ++A ++ ++++++Y+RLFG++V +NK + S ++G+LDI GFE+FE+NS E
Sbjct 418 KNSQQAKFILNALSRNLYERLFGYIVDMINKNLDHGSATLNYIGLLDIAGFEIFENNSFE 477
Query 63 QLLIN 67
QL IN
Sbjct 478 QLCIN 482
> Hs12667788
Length=1960
Score = 62.8 bits (151), Expect = 1e-10, Method: Composition-based stats.
Identities = 31/66 (46%), Positives = 44/66 (66%), Gaps = 1/66 (1%)
Query 3 RTHREADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNV-FMGMLDIFGFEVFEHNSL 61
+T +AD I+++AK Y+R+F WLV +NKA+ F+G+LDI GFE+F+ NS
Sbjct 405 QTKEQADFAIEALAKATYERMFRWLVLRINKALDKTKRQGASFIGILDIAGFEIFDLNSF 464
Query 62 EQLLIN 67
EQL IN
Sbjct 465 EQLCIN 470
> Hs19923615
Length=770
Score = 62.8 bits (151), Expect = 2e-10, Method: Composition-based stats.
Identities = 31/67 (46%), Positives = 40/67 (59%), Gaps = 1/67 (1%)
Query 2 PRTHREADMMIKSVAKHVYDRLFGWLVQFLNKAI-APPSGFNVFMGMLDIFGFEVFEHNS 60
P E D +AK +Y RLF WLV +N +I A + F+G+LD++GFE F NS
Sbjct 366 PCARAECDTRRDCLAKLIYARLFDWLVSVINSSICADTDSWTTFIGLLDVYGFESFPDNS 425
Query 61 LEQLLIN 67
LEQL IN
Sbjct 426 LEQLCIN 432
> Hs13124879
Length=1972
Score = 62.4 bits (150), Expect = 2e-10, Method: Composition-based stats.
Identities = 30/66 (45%), Positives = 44/66 (66%), Gaps = 1/66 (1%)
Query 3 RTHREADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNV-FMGMLDIFGFEVFEHNSL 61
+T +AD ++++AK Y+RLF W++ +NKA+ F+G+LDI GFE+FE NS
Sbjct 412 QTKEQADFAVEALAKATYERLFRWILTRVNKALDKTHRQGASFLGILDIAGFEIFEVNSF 471
Query 62 EQLLIN 67
EQL IN
Sbjct 472 EQLCIN 477
> Hs13124877
Length=1266
Score = 62.4 bits (150), Expect = 2e-10, Method: Composition-based stats.
Identities = 30/66 (45%), Positives = 44/66 (66%), Gaps = 1/66 (1%)
Query 3 RTHREADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNV-FMGMLDIFGFEVFEHNSL 61
+T +AD ++++AK Y+RLF W++ +NKA+ F+G+LDI GFE+FE NS
Sbjct 412 QTKEQADFAVEALAKATYERLFRWILTRVNKALDKTHRQGASFLGILDIAGFEIFEVNSF 471
Query 62 EQLLIN 67
EQL IN
Sbjct 472 EQLCIN 477
> Hs13124875
Length=1938
Score = 62.4 bits (150), Expect = 2e-10, Method: Composition-based stats.
Identities = 30/66 (45%), Positives = 44/66 (66%), Gaps = 1/66 (1%)
Query 3 RTHREADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNV-FMGMLDIFGFEVFEHNSL 61
+T +AD ++++AK Y+RLF W++ +NKA+ F+G+LDI GFE+FE NS
Sbjct 412 QTKEQADFAVEALAKATYERLFRWILTRVNKALDKTHRQGASFLGILDIAGFEIFEVNSF 471
Query 62 EQLLIN 67
EQL IN
Sbjct 472 EQLCIN 477
> Hs16160857
Length=1855
Score = 61.6 bits (148), Expect = 3e-10, Method: Composition-based stats.
Identities = 28/66 (42%), Positives = 42/66 (63%), Gaps = 0/66 (0%)
Query 2 PRTHREADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSL 61
P + +A ++AKH+Y +LF W+V +N+A+ + F+G+LDI+GFE FE NS
Sbjct 390 PISKLQATNARDALAKHIYAKLFNWIVDNVNQALHSAVKQHSFIGVLDIYGFETFEINSF 449
Query 62 EQLLIN 67
EQ IN
Sbjct 450 EQFCIN 455
> Hs10835119
Length=1855
Score = 61.6 bits (148), Expect = 3e-10, Method: Composition-based stats.
Identities = 28/66 (42%), Positives = 42/66 (63%), Gaps = 0/66 (0%)
Query 2 PRTHREADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSL 61
P + +A ++AKH+Y +LF W+V +N+A+ + F+G+LDI+GFE FE NS
Sbjct 390 PISKLQATNARDALAKHIYAKLFNWIVDNVNQALHSAVKQHSFIGVLDIYGFETFEINSF 449
Query 62 EQLLIN 67
EQ IN
Sbjct 450 EQFCIN 455
> 7297714
Length=1011
Score = 61.2 bits (147), Expect = 4e-10, Method: Composition-based stats.
Identities = 28/66 (42%), Positives = 43/66 (65%), Gaps = 5/66 (7%)
Query 7 EADMMIKSVAKHVYDRLFGWLVQFLNKAI-----APPSGFNVFMGMLDIFGFEVFEHNSL 61
+A+ ++AK +YDRLF W++ +N+AI + FN +G+LDI+GFE+F+ NS
Sbjct 331 QAEYGKDALAKAIYDRLFTWIISRINRAILFRGSKTQARFNSVIGVLDIYGFEIFDSNSF 390
Query 62 EQLLIN 67
EQ IN
Sbjct 391 EQFCIN 396
> Hs14772793
Length=1220
Score = 60.8 bits (146), Expect = 5e-10, Method: Composition-based stats.
Identities = 28/56 (50%), Positives = 37/56 (66%), Gaps = 0/56 (0%)
Query 12 IKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
+ ++AK YDRLF WLV +N+ + F+G+LDI GFE+FE NS EQL IN
Sbjct 125 VGALAKATYDRLFRWLVSRINQTLDTKLPRQFFIGVLDIAGFEIFEFNSFEQLCIN 180
> YKL129c
Length=1271
Score = 60.8 bits (146), Expect = 5e-10, Method: Composition-based stats.
Identities = 27/66 (40%), Positives = 43/66 (65%), Gaps = 0/66 (0%)
Query 2 PRTHREADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSL 61
P +A + ++AK +Y+ LF W+V +N ++ G + +G+LDI+GFE+FEHNS
Sbjct 361 PLNPVQATAVRDALAKAIYNNLFDWIVDRVNVSLQAFPGADKSIGILDIYGFEIFEHNSF 420
Query 62 EQLLIN 67
EQ+ IN
Sbjct 421 EQICIN 426
> CE27133
Length=2003
Score = 60.8 bits (146), Expect = 6e-10, Method: Composition-based stats.
Identities = 30/62 (48%), Positives = 42/62 (67%), Gaps = 1/62 (1%)
Query 7 EADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNV-FMGMLDIFGFEVFEHNSLEQLL 65
+ + + ++AK Y+RLF WLV LNK++ +V F+G+LDI GFE+FE NS EQL
Sbjct 429 QVNFSVGAIAKASYERLFRWLVHRLNKSLDRTRQQSVSFIGILDIAGFEIFETNSFEQLC 488
Query 66 IN 67
IN
Sbjct 489 IN 490
> Hs22065493
Length=1028
Score = 59.7 bits (143), Expect = 1e-09, Method: Composition-based stats.
Identities = 30/72 (41%), Positives = 41/72 (56%), Gaps = 6/72 (8%)
Query 2 PRTHREADMMIKSVAKHVYDRLFGWLVQFLNKAIA------PPSGFNVFMGMLDIFGFEV 55
P +A ++AK VY R F WLV +N+++A P +G+LDI+GFEV
Sbjct 333 PLNLEQAAYARDALAKAVYSRTFTWLVGKINRSLASKDVESPSWRSTTVLGLLDIYGFEV 392
Query 56 FEHNSLEQLLIN 67
F+HNS EQ IN
Sbjct 393 FQHNSFEQFCIN 404
> Hs7669506
Length=1939
Score = 59.3 bits (142), Expect = 2e-09, Method: Composition-based stats.
Identities = 26/56 (46%), Positives = 39/56 (69%), Gaps = 0/56 (0%)
Query 12 IKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
+ ++AK VYD++F W+V +N+ + F+G+LDI GFE+F+ NSLEQL IN
Sbjct 427 VGALAKAVYDKMFLWMVTRINQQLDTKQPRQYFIGVLDIAGFEIFDFNSLEQLCIN 482
> At1g08730
Length=1572
Score = 59.3 bits (142), Expect = 2e-09, Method: Composition-based stats.
Identities = 27/53 (50%), Positives = 35/53 (66%), Gaps = 0/53 (0%)
Query 15 VAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
+AK VY RLF WLV +NK+I + +G+LDI+GFE F+ NS EQ IN
Sbjct 408 LAKTVYSRLFDWLVDKINKSIGQDANSRSLIGVLDIYGFESFKTNSFEQFCIN 460
> Hs11342672
Length=1940
Score = 58.9 bits (141), Expect = 2e-09, Method: Composition-based stats.
Identities = 26/56 (46%), Positives = 40/56 (71%), Gaps = 0/56 (0%)
Query 12 IKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
+ +++K VY++LF W+V +N+ + F+G+LDI GFE+FE+NSLEQL IN
Sbjct 425 VNALSKSVYEKLFLWMVTRINQQLDTKLPRQHFIGVLDIAGFEIFEYNSLEQLCIN 480
> 7298338
Length=538
Score = 58.5 bits (140), Expect = 2e-09, Method: Composition-based stats.
Identities = 28/65 (43%), Positives = 40/65 (61%), Gaps = 0/65 (0%)
Query 3 RTHREADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLE 62
R ++ I ++ K V+DRLF WLV+ N+ + F+G+LDI GFE+FE+N E
Sbjct 414 RNVQQVTNSIGALCKGVFDRLFKWLVKKCNETLDTQQKRQHFIGVLDIAGFEIFEYNGFE 473
Query 63 QLLIN 67
QL IN
Sbjct 474 QLCIN 478
> YOR326w
Length=1574
Score = 58.5 bits (140), Expect = 3e-09, Method: Composition-based stats.
Identities = 28/57 (49%), Positives = 37/57 (64%), Gaps = 3/57 (5%)
Query 14 SVAKHVYDRLFGWLVQFLNKAIAPPS---GFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
SVAK +Y LF WLV+ +N + P+ + F+G+LDI+GFE FE NS EQ IN
Sbjct 410 SVAKFIYSALFDWLVENINTVLCNPAVNDQISSFIGVLDIYGFEHFEKNSFEQFCIN 466
> Hs4557773
Length=1935
Score = 58.2 bits (139), Expect = 3e-09, Method: Composition-based stats.
Identities = 25/54 (46%), Positives = 36/54 (66%), Gaps = 0/54 (0%)
Query 14 SVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
++AK VY+R+F W+V +N + F+G+LDI GFE+F+ NS EQL IN
Sbjct 426 ALAKAVYERMFNWMVTRINATLETKQPRQYFIGVLDIAGFEIFDFNSFEQLCIN 479
> CE28727
Length=1235
Score = 58.2 bits (139), Expect = 3e-09, Method: Composition-based stats.
Identities = 26/63 (41%), Positives = 43/63 (68%), Gaps = 1/63 (1%)
Query 6 READMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVF-MGMLDIFGFEVFEHNSLEQL 64
++ D + +++K +Y R+F WL++ +NK + S V+ +G+LDI GFE+F+ NS EQL
Sbjct 411 QQVDWAVGALSKAIYARMFNWLIKRVNKTLQANSDEMVYYIGVLDIAGFEIFDRNSFEQL 470
Query 65 LIN 67
IN
Sbjct 471 WIN 473
> 7291892
Length=2056
Score = 58.2 bits (139), Expect = 4e-09, Method: Composition-based stats.
Identities = 26/66 (39%), Positives = 43/66 (65%), Gaps = 1/66 (1%)
Query 3 RTHREADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNV-FMGMLDIFGFEVFEHNSL 61
+T + + ++++AK Y+R+F WLV +N+++ F+G+LD+ GFE+FE NS
Sbjct 503 QTKEQVEFAVEAIAKACYERMFKWLVNRINRSLDRTKRQGASFIGILDMAGFEIFELNSF 562
Query 62 EQLLIN 67
EQL IN
Sbjct 563 EQLCIN 568
> Hs20542063
Length=1939
Score = 57.8 bits (138), Expect = 4e-09, Method: Composition-based stats.
Identities = 25/56 (44%), Positives = 37/56 (66%), Gaps = 0/56 (0%)
Query 12 IKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
I ++AK VY+++F W+V +N + F+G+LDI GFE+F+ NS EQL IN
Sbjct 425 IGALAKAVYEKMFNWMVTRINATLETKQPRQYFIGVLDIAGFEIFDFNSFEQLCIN 480
> Hs8923940
Length=1941
Score = 57.8 bits (138), Expect = 5e-09, Method: Composition-based stats.
Identities = 26/65 (40%), Positives = 42/65 (64%), Gaps = 0/65 (0%)
Query 3 RTHREADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLE 62
+T + + ++AK VY+++F W+V +N+ + F+G+LDI GFE+F+ NSLE
Sbjct 418 QTVEQVSNAVGALAKAVYEKMFLWMVARINQQLDTKQPRQYFIGVLDIAGFEIFDFNSLE 477
Query 63 QLLIN 67
QL IN
Sbjct 478 QLCIN 482
> CE04618
Length=1956
Score = 57.8 bits (138), Expect = 5e-09, Method: Composition-based stats.
Identities = 27/66 (40%), Positives = 43/66 (65%), Gaps = 1/66 (1%)
Query 3 RTHREADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNV-FMGMLDIFGFEVFEHNSL 61
+ +A+ ++++AK Y+RLF WLV +NK++ F+G+LDI GFE+F+ NS
Sbjct 412 QNQEQAEFAVEAIAKASYERLFKWLVTRINKSLDRTHRQGASFIGILDIAGFEIFDINSF 471
Query 62 EQLLIN 67
EQ+ IN
Sbjct 472 EQICIN 477
> Hs4505301
Length=1937
Score = 57.8 bits (138), Expect = 5e-09, Method: Composition-based stats.
Identities = 25/56 (44%), Positives = 39/56 (69%), Gaps = 0/56 (0%)
Query 12 IKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
+ ++AK VY+++F W+V +N+ + F+G+LDI GFE+F+ NSLEQL IN
Sbjct 427 VGALAKAVYEKMFLWMVTRINQQLDTKQPRQYFIGVLDIAGFEIFDFNSLEQLCIN 482
> Hs20558376
Length=1937
Score = 57.8 bits (138), Expect = 5e-09, Method: Composition-based stats.
Identities = 25/56 (44%), Positives = 39/56 (69%), Gaps = 0/56 (0%)
Query 12 IKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
+ ++AK VY+++F W+V +N+ + F+G+LDI GFE+F+ NSLEQL IN
Sbjct 427 VGALAKAVYEKMFLWMVTRINQQLDTKQPRQYFIGVLDIAGFEIFDFNSLEQLCIN 482
> Hs11321579
Length=1938
Score = 57.8 bits (138), Expect = 5e-09, Method: Composition-based stats.
Identities = 25/56 (44%), Positives = 39/56 (69%), Gaps = 0/56 (0%)
Query 12 IKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
+ ++AK VY+++F W+V +N+ + F+G+LDI GFE+F+ NSLEQL IN
Sbjct 426 VGALAKAVYEKMFLWMVTRINQQLDTKQPRQYFIGVLDIAGFEIFDFNSLEQLCIN 481
> Hs22064265
Length=1006
Score = 57.4 bits (137), Expect = 6e-09, Method: Composition-based stats.
Identities = 29/70 (41%), Positives = 42/70 (60%), Gaps = 6/70 (8%)
Query 4 THREADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPS------GFNVFMGMLDIFGFEVFE 57
T +EA + AK +Y+RLF W+V +N I + G N +G+LDI+GFE+F+
Sbjct 332 TEQEASYGRDAFAKAIYERLFCWIVTRINDIIEVKNYDTTIHGKNTVIGVLDIYGFEIFD 391
Query 58 HNSLEQLLIN 67
+NS EQ IN
Sbjct 392 NNSFEQFCIN 401
> Hs11024712
Length=1939
Score = 57.0 bits (136), Expect = 8e-09, Method: Composition-based stats.
Identities = 24/56 (42%), Positives = 39/56 (69%), Gaps = 0/56 (0%)
Query 12 IKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
+ ++AK +Y+++F W+V +N+ + F+G+LDI GFE+F+ NSLEQL IN
Sbjct 427 VGALAKAIYEKMFLWMVTRINQQLDTKQPRQYFIGVLDIAGFEIFDFNSLEQLCIN 482
> At2g33240
Length=1611
Score = 57.0 bits (136), Expect = 9e-09, Method: Composition-based stats.
Identities = 26/54 (48%), Positives = 35/54 (64%), Gaps = 0/54 (0%)
Query 14 SVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
++AK VY +LF WLV +N +I S +G+LDI+GFE F+ NS EQ IN
Sbjct 413 ALAKIVYSKLFDWLVTKINNSIGQDSSSKYIIGVLDIYGFESFKTNSFEQFCIN 466
> At2g31900
Length=1490
Score = 56.6 bits (135), Expect = 1e-08, Method: Composition-based stats.
Identities = 26/54 (48%), Positives = 35/54 (64%), Gaps = 0/54 (0%)
Query 14 SVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
++AK VY RLF W+V +N +I +G+LDI+GFE F+ NS EQL IN
Sbjct 333 ALAKTVYSRLFDWIVDKINSSIGQDPDAKSLIGVLDIYGFESFKINSFEQLCIN 386
> At4g33200
Length=1374
Score = 56.6 bits (135), Expect = 1e-08, Method: Composition-based stats.
Identities = 28/56 (50%), Positives = 38/56 (67%), Gaps = 4/56 (7%)
Query 14 SVAKHVYDRLFGWLVQFLNKAIA--PPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
++AK VY LF WLV +NK++ P S F + G+LDI+GFE F++NS EQ IN
Sbjct 396 TLAKTVYAHLFDWLVDKINKSVGQDPESRFQI--GVLDIYGFECFKNNSFEQFCIN 449
> 7304206
Length=1786
Score = 56.2 bits (134), Expect = 1e-08, Method: Composition-based stats.
Identities = 26/51 (50%), Positives = 35/51 (68%), Gaps = 0/51 (0%)
Query 17 KHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
KH+Y +LF ++V LNK++ S F+G+LDI+GFE FE NS EQ IN
Sbjct 415 KHIYAKLFQYIVGVLNKSLNNGSKQCSFIGVLDIYGFETFEVNSFEQFCIN 465
> At1g54560
Length=1529
Score = 56.2 bits (134), Expect = 1e-08, Method: Composition-based stats.
Identities = 25/53 (47%), Positives = 35/53 (66%), Gaps = 0/53 (0%)
Query 15 VAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
+AK +Y RLF WLV+ +N +I + +G+LDI+GFE F+ NS EQ IN
Sbjct 403 LAKTIYSRLFDWLVEKINVSIGQDATSRSLIGVLDIYGFESFKTNSFEQFCIN 455
> Hs8393810_2
Length=1279
Score = 56.2 bits (134), Expect = 1e-08, Method: Composition-based stats.
Identities = 31/71 (43%), Positives = 44/71 (61%), Gaps = 5/71 (7%)
Query 2 PRTHREADMMIKSVAKHVYDRLFGWLVQFLNKAI---APPSGF--NVFMGMLDIFGFEVF 56
P T +A + ++AK +Y RLF W+V +N + + PSG + +G+LDIFGFE F
Sbjct 329 PNTVEKATDVRDAMAKTLYGRLFSWIVNCINSLLKHDSSPSGNGDELSIGILDIFGFENF 388
Query 57 EHNSLEQLLIN 67
+ NS EQL IN
Sbjct 389 KKNSFEQLCIN 399
> Hs20544910_2
Length=1280
Score = 56.2 bits (134), Expect = 1e-08, Method: Composition-based stats.
Identities = 31/71 (43%), Positives = 44/71 (61%), Gaps = 5/71 (7%)
Query 2 PRTHREADMMIKSVAKHVYDRLFGWLVQFLNKAI---APPSGF--NVFMGMLDIFGFEVF 56
P T +A + ++AK +Y RLF W+V +N + + PSG + +G+LDIFGFE F
Sbjct 329 PNTVEKATDVRDAMAKTLYGRLFSWIVNCINSLLKHDSSPSGNGDELSIGILDIFGFENF 388
Query 57 EHNSLEQLLIN 67
+ NS EQL IN
Sbjct 389 KKNSFEQLCIN 399
> Hs4885503
Length=1043
Score = 56.2 bits (134), Expect = 1e-08, Method: Composition-based stats.
Identities = 26/55 (47%), Positives = 38/55 (69%), Gaps = 1/55 (1%)
Query 14 SVAKHVYDRLFGWLVQFLNKAIAPPSG-FNVFMGMLDIFGFEVFEHNSLEQLLIN 67
++AK++Y RLF W+V +N++I G MG+LDI+GFE+ E NS EQ +IN
Sbjct 344 ALAKNIYSRLFDWIVNRINESIKVGIGEKKKVMGVLDIYGFEILEDNSFEQFVIN 398
> YAL029c
Length=1471
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 29/59 (49%), Positives = 35/59 (59%), Gaps = 5/59 (8%)
Query 14 SVAKHVYDRLFGWLVQFLNKAIAPPS-----GFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
SVAK +Y LF WLV +NK + P F+G+LDI+GFE FE NS EQ IN
Sbjct 409 SVAKFIYSTLFDWLVDNINKTLYDPELDQQDHVFSFIGILDIYGFEHFEKNSFEQFCIN 467
> At3g58160
Length=1242
Score = 55.8 bits (133), Expect = 2e-08, Method: Composition-based stats.
Identities = 26/53 (49%), Positives = 34/53 (64%), Gaps = 0/53 (0%)
Query 15 VAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
+AK +Y RLF WLV +N +I S +G+LDI+GFE F+ NS EQ IN
Sbjct 398 LAKTIYSRLFDWLVNKINISIGQDSHSRRLIGVLDIYGFESFKTNSFEQFCIN 450
> At4g28710
Length=899
Score = 55.8 bits (133), Expect = 2e-08, Method: Composition-based stats.
Identities = 25/54 (46%), Positives = 34/54 (62%), Gaps = 0/54 (0%)
Query 14 SVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
++AK VY RLF W+V +N +I +G+LDI+GFE F+ NS EQ IN
Sbjct 398 ALAKTVYSRLFDWIVNKINDSIGQDPDSEYLIGVLDIYGFESFKTNSFEQFCIN 451
> At1g04160
Length=1519
Score = 55.8 bits (133), Expect = 2e-08, Method: Composition-based stats.
Identities = 25/54 (46%), Positives = 35/54 (64%), Gaps = 0/54 (0%)
Query 14 SVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
++AK +Y RLF WLV+ +N +I +G+LDI+GFE F+ NS EQ IN
Sbjct 400 ALAKVMYSRLFDWLVEKINTSIGQDPDSKYLIGVLDIYGFESFKTNSFEQFCIN 453
> Hs22062833
Length=930
Score = 55.5 bits (132), Expect = 2e-08, Method: Composition-based stats.
Identities = 30/70 (42%), Positives = 38/70 (54%), Gaps = 6/70 (8%)
Query 4 THREADMMIKSVAKHVYDRLFGWLVQFLNKAIAPP------SGFNVFMGMLDIFGFEVFE 57
T EA + AK VY RLF W+V +N + P G + +G+LDI+GFEVF
Sbjct 256 TAAEASYARDACAKAVYQRLFEWVVNRINSVMEPRGRDPRRDGKDTVIGVLDIYGFEVFP 315
Query 58 HNSLEQLLIN 67
NS EQ IN
Sbjct 316 VNSFEQFCIN 325
> Hs20428779_2
Length=921
Score = 55.5 bits (132), Expect = 2e-08, Method: Composition-based stats.
Identities = 29/67 (43%), Positives = 42/67 (62%), Gaps = 6/67 (8%)
Query 6 READMMIKSVAKHVYDRLFGWLVQFLNKAIAP-----PSGFNVFMGMLDIFGFEVFEHNS 60
R AD+ +++K +Y RLF W+V +N + P +G + +G+LDIFGFE F+ NS
Sbjct 318 RAADVR-DAMSKALYGRLFSWIVNRINTLLQPDENICSAGGGMNVGILDIFGFENFQRNS 376
Query 61 LEQLLIN 67
EQL IN
Sbjct 377 FEQLCIN 383
> At2g20290
Length=1502
Score = 55.1 bits (131), Expect = 3e-08, Method: Composition-based stats.
Identities = 25/54 (46%), Positives = 34/54 (62%), Gaps = 0/54 (0%)
Query 14 SVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
++AK VY RLF W+V +N +I +G+LDI+GFE F+ NS EQ IN
Sbjct 408 ALAKFVYSRLFDWIVNKINNSIGQDPDSKDMIGVLDIYGFESFKTNSFEQFCIN 461
> At5g43900
Length=1505
Score = 55.1 bits (131), Expect = 3e-08, Method: Composition-based stats.
Identities = 25/54 (46%), Positives = 34/54 (62%), Gaps = 0/54 (0%)
Query 14 SVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLEQLLIN 67
++AK +Y RLF WLV +N +I +G+LDI+GFE F+ NS EQ IN
Sbjct 399 ALAKVMYSRLFDWLVDKINSSIGQDHDSKYLIGVLDIYGFESFKTNSFEQFCIN 452
> ECU04g1000
Length=1700
Score = 54.3 bits (129), Expect = 5e-08, Method: Composition-based stats.
Identities = 25/65 (38%), Positives = 43/65 (66%), Gaps = 1/65 (1%)
Query 3 RTHREADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSLE 62
R+ +A ++ +++ +YD++F ++ +N ++ P N F+G+LDI GFE+FE NS E
Sbjct 423 RSREQALKIVDGLSRILYDKMFEGVIDRINMSLDSPHKGN-FIGVLDIAGFEIFEKNSFE 481
Query 63 QLLIN 67
QL IN
Sbjct 482 QLCIN 486
> Hs9055284
Length=1742
Score = 54.3 bits (129), Expect = 6e-08, Method: Composition-based stats.
Identities = 26/66 (39%), Positives = 41/66 (62%), Gaps = 0/66 (0%)
Query 2 PRTHREADMMIKSVAKHVYDRLFGWLVQFLNKAIAPPSGFNVFMGMLDIFGFEVFEHNSL 61
P T +A ++AK +Y LF ++V+ +N+A+ + F+G+LDI+GFE F+ NS
Sbjct 388 PMTRPQAVNARDALAKKIYAHLFDFIVERINQALQFSGKQHTFIGVLDIYGFETFDVNSF 447
Query 62 EQLLIN 67
EQ IN
Sbjct 448 EQFCIN 453
Lambda K H
0.328 0.143 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160559522
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40