bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0469_orf2
Length=185
Score E
Sequences producing significant alignments: (Bits) Value
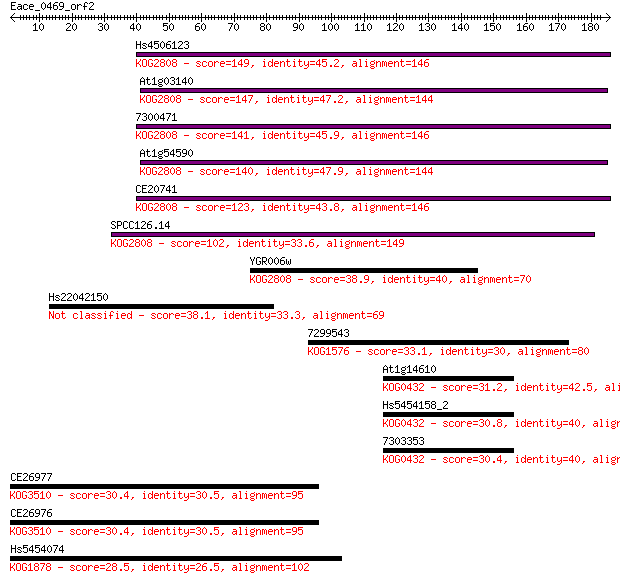
Hs4506123 149 3e-36
At1g03140 147 1e-35
7300471 141 9e-34
At1g54590 140 1e-33
CE20741 123 2e-28
SPCC126.14 102 4e-22
YGR006w 38.9 0.007
Hs22042150 38.1 0.011
7299543 33.1 0.37
At1g14610 31.2 1.2
Hs5454158_2 30.8 1.6
7303353 30.4 2.0
CE26977 30.4 2.2
CE26976 30.4 2.2
Hs5454074 28.5 7.3
> Hs4506123
Length=342
Score = 149 bits (376), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 66/146 (45%), Positives = 99/146 (67%), Gaps = 0/146 (0%)
Query 40 VVIAWARKMLSLWEDELKHRSEDEKATAEGRQQTALHRQTKKDLKPLFKKLKQRDLEADI 99
++ + + +L +W EL R + K + +G+ +A +QT+ L+PLF+KL++R+L ADI
Sbjct 187 IITKFLKFLLGVWAKELNAREDYVKRSVQGKLNSATQKQTESYLRPLFRKLRKRNLPADI 246
Query 100 LEKLFSIVQLCDERRYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHIL 159
E + I++ +R Y A+ A++ +AIGNA WP+GVTMVGIH R GR K+ + VAH+L
Sbjct 247 KESITDIIKFMLQREYVKANDAYLQMAIGNAPWPIGVTMVGIHARTGREKIFSKHVAHVL 306
Query 160 NDETTRKYIQMFKRLMSFAQRKFPAN 185
NDET RKYIQ KRLM+ Q+ FP +
Sbjct 307 NDETQRKYIQGLKRLMTICQKHFPTD 332
> At1g03140
Length=420
Score = 147 bits (371), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 68/144 (47%), Positives = 96/144 (66%), Gaps = 0/144 (0%)
Query 41 VIAWARKMLSLWEDELKHRSEDEKATAEGRQQTALHRQTKKDLKPLFKKLKQRDLEADIL 100
++ + +K+L W+ EL E+ TA+G+Q A +Q + L PLF +++ L ADI
Sbjct 229 ILVFYKKLLIEWKQELDAMENTERRTAKGKQMVATFKQCARYLVPLFNLCRKKGLPADIR 288
Query 101 EKLFSIVQLCDERRYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHILN 160
+ L +V C +R Y A ++ LAIGNA WP+GVTMVGIHER+ R K+ T+ VAHI+N
Sbjct 289 QALMVMVNHCIKRDYLAAMDHYIKLAIGNAPWPIGVTMVGIHERSAREKIYTNSVAHIMN 348
Query 161 DETTRKYIQMFKRLMSFAQRKFPA 184
DETTRKY+Q KRLM+F QR++P
Sbjct 349 DETTRKYLQSVKRLMTFCQRRYPT 372
> 7300471
Length=340
Score = 141 bits (355), Expect = 9e-34, Method: Compositional matrix adjust.
Identities = 67/146 (45%), Positives = 93/146 (63%), Gaps = 0/146 (0%)
Query 40 VVIAWARKMLSLWEDELKHRSEDEKATAEGRQQTALHRQTKKDLKPLFKKLKQRDLEADI 99
V+I +L LW D++ + S+ EK + + + ++ QTK+ +KPLF+KLK L DI
Sbjct 181 VIITLLTFLLKLWNDQIANYSKHEKMSTKVKMTRVIYTQTKEYVKPLFRKLKHHTLPEDI 240
Query 100 LEKLFSIVQLCDERRYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHIL 159
L+ L I + R Y A A++ +AIGNA WP+GVTMVGIH R GR K+ + VAH++
Sbjct 241 LDSLRDICKHLLNRNYITASDAYLEMAIGNAPWPIGVTMVGIHARTGREKIFSKNVAHVM 300
Query 160 NDETTRKYIQMFKRLMSFAQRKFPAN 185
NDET RKYIQ KRLM+ Q FP +
Sbjct 301 NDETQRKYIQGLKRLMTKCQEYFPTD 326
> At1g54590
Length=256
Score = 140 bits (354), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 69/145 (47%), Positives = 95/145 (65%), Gaps = 1/145 (0%)
Query 41 VIAWARKMLSLWEDELKHRSEDEKATAEGRQQTALHRQTKKDLKPLFKKLKQRDLEADIL 100
++ + +K+L W+ EL+ E+ TA G+Q A Q + L PLF + + L ADI
Sbjct 64 ILVFCKKLLLEWKQELEAMENTERRTAIGKQMLATFNQCARYLTPLFHLCRNKCLPADIR 123
Query 101 EKLFSIVQLCDERRYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNT-SQVAHIL 159
+ L +V +R Y DA F+ LAIGNA WP+GVTMVGIHER+ R K++T S VAHI+
Sbjct 124 QGLMVMVNCWIKRDYLDATAQFIKLAIGNAPWPIGVTMVGIHERSAREKISTSSSVAHIM 183
Query 160 NDETTRKYIQMFKRLMSFAQRKFPA 184
N+ETTRKY+Q KRLM+F QR++ A
Sbjct 184 NNETTRKYLQSVKRLMTFCQRRYSA 208
> CE20741
Length=348
Score = 123 bits (309), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 64/147 (43%), Positives = 89/147 (60%), Gaps = 5/147 (3%)
Query 40 VVIAWARKMLSLWEDELKHRSEDEKATAEGRQQTALHRQTKKDLKPLFKKLKQRDLEADI 99
++++ R +L+ W +L R D K TA Q A H+QT LK L +++ + DI
Sbjct 194 IILSICRYILARWAKDLNDRPLDVKKTA----QAAHHKQTMMHLKSLMTSMERYNCNNDI 249
Query 100 LEKLFSIVQL-CDERRYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHI 158
L I +L +R Y +A+ A+M +AIGNA WP+GVT GIH+R G +K S +AH+
Sbjct 250 RHHLAKICRLLVIDRNYLEANNAYMEMAIGNAPWPVGVTRSGIHQRPGSAKSYVSNIAHV 309
Query 159 LNDETTRKYIQMFKRLMSFAQRKFPAN 185
LNDET RKYIQ FKRLM+ Q FP +
Sbjct 310 LNDETQRKYIQAFKRLMTKMQEYFPTD 336
> SPCC126.14
Length=343
Score = 102 bits (254), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 50/149 (33%), Positives = 84/149 (56%), Gaps = 0/149 (0%)
Query 32 TTEKSKEAVVIAWARKMLSLWEDELKHRSEDEKATAEGRQQTALHRQTKKDLKPLFKKLK 91
TT+ V+A+ + + +W++ L +S + ++E + Q + RQ K+DL L + +
Sbjct 185 TTKPKVSKQVVAFLQHGIRIWDNFLSSKSINSFESSESQMQLKIFRQAKQDLDVLIQLIV 244
Query 92 QRDLEADILEKLFSIVQLCDERRYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLN 151
L DI + + I C + + A+ ++ L IGNA WP+GVTMVGIHER+ +L
Sbjct 245 DEALNDDIFKSIAEICYRCQKHEFVKANDMYLRLTIGNAPWPIGVTMVGIHERSAHQRLQ 304
Query 152 TSQVAHILNDETTRKYIQMFKRLMSFAQR 180
+ ++IL DE RK +Q KR ++F +R
Sbjct 305 ANPSSNILKDEKKRKCLQALKRFITFQER 333
> YGR006w
Length=219
Score = 38.9 bits (89), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 28/75 (37%), Positives = 42/75 (56%), Gaps = 5/75 (6%)
Query 75 LHRQTKKDLKPLFKKLKQRDLEADILEKLFSIV-QLCDERRYRDAHGAFMLLAIGNAAWP 133
L TKK L PL +L++ L D+L L +++ L + A ++M L+IGN AWP
Sbjct 135 LFLDTKKALFPLLLQLRRNQLAPDLLISLATVLYHLQQPKEINLAVQSYMKLSIGNVAWP 194
Query 134 MGVT----MVGIHER 144
+GVT M+ +H R
Sbjct 195 IGVTSVAFMLVVHIR 209
> Hs22042150
Length=1717
Score = 38.1 bits (87), Expect = 0.011, Method: Composition-based stats.
Identities = 23/69 (33%), Positives = 38/69 (55%), Gaps = 4/69 (5%)
Query 13 GKKETNSESTDAKQSQNEETTEKSKEAVVIAWARKMLSLWEDELKHRSEDEKATAEGRQQ 72
G + SE TD K S+N E +K++ ++ R++ + WE L RS E++ A G QQ
Sbjct 1191 GSHQQASE-TDTKTSENPEEEVSTKDSAILR--RRVPAGWE-RLHVRSTQEQSAAAGEQQ 1246
Query 73 TALHRQTKK 81
LH++ +
Sbjct 1247 QLLHQERSR 1255
> 7299543
Length=345
Score = 33.1 bits (74), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 37/80 (46%), Gaps = 9/80 (11%)
Query 93 RDLEADILEKLFSIVQLCDERRYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNT 152
+DL+ I E L ++ QL E + R IG +A+P+ V + AGR
Sbjct 149 KDLDIVINETLPTLEQLVKEGKAR---------FIGVSAYPISVLKEFLTRTAGRLDTVL 199
Query 153 SQVAHILNDETTRKYIQMFK 172
+ + L DET +Y+ FK
Sbjct 200 TYARYTLTDETLLEYLDFFK 219
> At1g14610
Length=1108
Score = 31.2 bits (69), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 25/44 (56%), Gaps = 4/44 (9%)
Query 116 RDAHGAFMLLAIGNAAWPM----GVTMVGIHERAGRSKLNTSQV 155
RDAHG M ++GN P+ GVT+ G+H+R L+ +V
Sbjct 689 RDAHGRKMSKSLGNVIDPLEVINGVTLEGLHKRLEEGNLDPKEV 732
> Hs5454158_2
Length=985
Score = 30.8 bits (68), Expect = 1.6, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 26/44 (59%), Gaps = 4/44 (9%)
Query 116 RDAHGAFMLLAIGNAAWPM----GVTMVGIHERAGRSKLNTSQV 155
RDAHG M ++GN P+ G+++ G+H + S L+ S+V
Sbjct 577 RDAHGRKMSKSLGNVIDPLDVIYGISLQGLHNQLLNSNLDPSEV 620
> 7303353
Length=1049
Score = 30.4 bits (67), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 25/44 (56%), Gaps = 4/44 (9%)
Query 116 RDAHGAFMLLAIGNAAWPM----GVTMVGIHERAGRSKLNTSQV 155
RDAHG M ++GN PM G+T+ G+H + S L+ ++
Sbjct 648 RDAHGRKMSKSLGNVIDPMDVIRGITLEGLHAQLVGSNLDPREI 691
> CE26977
Length=4450
Score = 30.4 bits (67), Expect = 2.2, Method: Composition-based stats.
Identities = 29/115 (25%), Positives = 47/115 (40%), Gaps = 24/115 (20%)
Query 1 EEEEEFDEDANK-----GKKETNSESTDAKQSQN-------EETTEKSKEAVVIAWARKM 48
+ E+E+D + G E + E T Q QN E T+K+ ++ + W
Sbjct 2794 DPEQEYDIQVSAHTEGGGWSEWSDELTSRTQQQNIPVLERELEVTDKTSNSISLKWE--- 2850
Query 49 LSLWEDELKH--------RSEDEKATAEGRQQTALHRQTKKDLKPLFKKLKQRDL 95
L +D+ H +SED+ A + HR+T D K K+L+ L
Sbjct 2851 -GLPQDQATHVVGYVLEFKSEDDNAEWQEYNGVVKHRKTSSDYKITVKQLETATL 2904
> CE26976
Length=4280
Score = 30.4 bits (67), Expect = 2.2, Method: Composition-based stats.
Identities = 29/115 (25%), Positives = 47/115 (40%), Gaps = 24/115 (20%)
Query 1 EEEEEFDEDANK-----GKKETNSESTDAKQSQN-------EETTEKSKEAVVIAWARKM 48
+ E+E+D + G E + E T Q QN E T+K+ ++ + W
Sbjct 2794 DPEQEYDIQVSAHTEGGGWSEWSDELTSRTQQQNIPVLERELEVTDKTSNSISLKWE--- 2850
Query 49 LSLWEDELKH--------RSEDEKATAEGRQQTALHRQTKKDLKPLFKKLKQRDL 95
L +D+ H +SED+ A + HR+T D K K+L+ L
Sbjct 2851 -GLPQDQATHVVGYVLEFKSEDDNAEWQEYNGVVKHRKTSSDYKITVKQLETATL 2904
> Hs5454074
Length=2517
Score = 28.5 bits (62), Expect = 7.3, Method: Composition-based stats.
Identities = 27/113 (23%), Positives = 55/113 (48%), Gaps = 11/113 (9%)
Query 1 EEEEEFDEDANKGKKETNSES-TDAKQSQNEETTEKSKEAVVIAWARKMLSLWEDELKHR 59
E+ +E + A+KG+K NS+ + +++ S+EA+ + ++ S+ +E
Sbjct 556 EDNDEKEAVASKGRKTANSQGRRKGRITRSMANEANSEEAITPQQSAELASMELNESSRW 615
Query 60 SEDEKATAE------GRQQTALHR----QTKKDLKPLFKKLKQRDLEADILEK 102
+E+E TA+ GR +A+ R +T K + K+R +IL++
Sbjct 616 TEEEMETAKKGLLEHGRNWSAIARMVGSKTVSQCKNFYFNYKKRQNLDEILQQ 668
Lambda K H
0.313 0.126 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2986559618
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40