bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0463_orf1
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
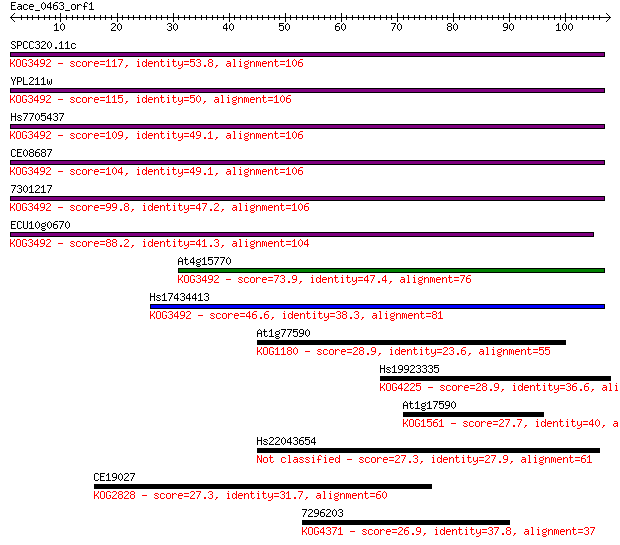
SPCC320.11c 117 4e-27
YPL211w 115 3e-26
Hs7705437 109 1e-24
CE08687 104 4e-23
7301217 99.8 1e-21
ECU10g0670 88.2 4e-18
At4g15770 73.9 7e-14
Hs17434413 46.6 1e-05
At1g77590 28.9 2.6
Hs19923335 28.9 2.7
At1g17590 27.7 5.6
Hs22043654 27.3 6.2
CE19027 27.3 7.9
7296203 26.9 9.8
> SPCC320.11c
Length=180
Score = 117 bits (294), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 57/106 (53%), Positives = 72/106 (67%), Gaps = 1/106 (0%)
Query 1 KGRKFRLGITALHLIARYAPHKVWLNPGGEQAFVYGNHIVKRHVGRLTADMPSGAGVCVL 60
K KFRL ITAL IA+YA +K+W+ GE F+YGNH++K HVGR+T D P GV +
Sbjct 75 KTNKFRLHITALDYIAQYARYKIWVKSNGEMPFLYGNHVLKAHVGRITDDTPQHQGVVIY 134
Query 61 SLNGDLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEADL 106
S+N D PLGFG +A+ST E+ AI +H ADVGEYLR+E L
Sbjct 135 SMN-DTPLGFGVTARSTLELRRLEPTAIVAFHQADVGEYLRDEDTL 179
> YPL211w
Length=181
Score = 115 bits (287), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 53/106 (50%), Positives = 75/106 (70%), Gaps = 1/106 (0%)
Query 1 KGRKFRLGITALHLIARYAPHKVWLNPGGEQAFVYGNHIVKRHVGRLTADMPSGAGVCVL 60
K KFRL IT+L ++A++A +K+W+ P GE F+YGNH++K HVG+++ D+P AGV V
Sbjct 75 KTGKFRLHITSLTVLAKHAKYKIWIKPNGEMPFLYGNHVLKAHVGKMSDDIPEHAGVIVF 134
Query 61 SLNGDLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEADL 106
++N D+PLGFG SAKST+E I + AD+GEYLR+E L
Sbjct 135 AMN-DVPLGFGVSAKSTSESRNMQPTGIVAFRQADIGEYLRDEDTL 179
> Hs7705437
Length=180
Score = 109 bits (272), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 52/106 (49%), Positives = 70/106 (66%), Gaps = 1/106 (0%)
Query 1 KGRKFRLGITALHLIARYAPHKVWLNPGGEQAFVYGNHIVKRHVGRLTADMPSGAGVCVL 60
K KFRL +TAL +A YA +KVW+ PG EQ+F+YGNH++K +GR+T + GV V
Sbjct 75 KTHKFRLHVTALDYLAPYAKYKVWIKPGAEQSFLYGNHVLKSGLGRITENTSQYQGVVVY 134
Query 61 SLNGDLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEADL 106
S+ D+PLGFG +AKST + AI +H AD+GEY+R E L
Sbjct 135 SM-ADIPLGFGVAAKSTQDCRKVDPMAIVVFHQADIGEYVRHEETL 179
> CE08687
Length=180
Score = 104 bits (260), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 52/106 (49%), Positives = 68/106 (64%), Gaps = 1/106 (0%)
Query 1 KGRKFRLGITALHLIARYAPHKVWLNPGGEQAFVYGNHIVKRHVGRLTADMPSGAGVCVL 60
K +KF L ITAL +A YA KVWL P EQ F+YGN+I+K + R+T P+ AG+ V
Sbjct 75 KSKKFHLQITALDYLAPYAKFKVWLKPNAEQQFLYGNNILKSGIARMTDGTPTHAGIVVY 134
Query 61 SLNGDLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEADL 106
S+ D+PLGFG SAK T++ A A+ H D+GEYLR E+ L
Sbjct 135 SMT-DVPLGFGVSAKGTSDSKRADPTALVVLHQCDLGEYLRNESHL 179
> 7301217
Length=180
Score = 99.8 bits (247), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 50/106 (47%), Positives = 66/106 (62%), Gaps = 1/106 (0%)
Query 1 KGRKFRLGITALHLIARYAPHKVWLNPGGEQAFVYGNHIVKRHVGRLTADMPSGAGVCVL 60
K K + ITAL+ +A YA +KVW+ P EQ F+YGNHI K +GR+T + GV V
Sbjct 75 KTNKLKFHITALYYLAPYAQYKVWVKPSFEQQFLYGNHIPKTGLGRITENAGQYQGVVVY 134
Query 61 SLNGDLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEADL 106
S+N DLPLGFG A+ST + TA +H +D+GEY+R E L
Sbjct 135 SMN-DLPLGFGVLARSTTDCKTADPMTTVCFHQSDIGEYIRAEDTL 179
> ECU10g0670
Length=176
Score = 88.2 bits (217), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 43/104 (41%), Positives = 65/104 (62%), Gaps = 1/104 (0%)
Query 1 KGRKFRLGITALHLIARYAPHKVWLNPGGEQAFVYGNHIVKRHVGRLTADMPSGAGVCVL 60
K FRLG+ +L+L++R+A HKVW+ E +VYGN+ +K HV R++ +P +GV V
Sbjct 72 KSGSFRLGVASLNLLSRWAIHKVWIKNSAEMNYVYGNNALKSHVFRISEGIPINSGVFVF 131
Query 61 SLNGDLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEA 104
+ + D+PLGFG +A S A A++ +D GEY+R EA
Sbjct 132 NQH-DVPLGFGITALSPQGYSAAKGHALAILRQSDTGEYVRNEA 174
> At4g15770
Length=75
Score = 73.9 bits (180), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 36/76 (47%), Positives = 49/76 (64%), Gaps = 1/76 (1%)
Query 31 QAFVYGNHIVKRHVGRLTADMPSGAGVCVLSLNGDLPLGFGTSAKSTAEIHTAPTEAISF 90
+F+YGNH++K +GR+T + G GV V S++ D+PLGFG +AKST + I
Sbjct 1 MSFLYGNHVLKGGLGRITDSIVPGDGVVVFSMS-DVPLGFGIAAKSTQDCRKLDPNGIVV 59
Query 91 YHHADVGEYLREEADL 106
H AD+GEYLR E DL
Sbjct 60 LHQADIGEYLRGEDDL 75
> Hs17434413
Length=133
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 31/84 (36%), Positives = 40/84 (47%), Gaps = 11/84 (13%)
Query 26 NPGGEQAFVYGN---HIVKRHVGRLTADMPSGAGVCVLSLNGDLPLGFGTSAKSTAEIHT 82
N G++ G + K H R T GV V S+ D+PLGFG +AKST +
Sbjct 57 NISGDKLVSLGTCFGKVTKTHKFRFT-------GVVVYSM-ADIPLGFGVAAKSTQDCRK 108
Query 83 APTEAISFYHHADVGEYLREEADL 106
AI H AD+GEY + E L
Sbjct 109 VDPMAIVVLHQADIGEYAQHEETL 132
> At1g77590
Length=691
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 13/55 (23%), Positives = 29/55 (52%), Gaps = 1/55 (1%)
Query 45 GRLTADMPSGAGVCVLSLNGDLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEY 99
G LT+D P G V+ ++ LG+ + + T E++ + + +++ D+G +
Sbjct 487 GYLTSDKPMPRGEIVIG-GSNITLGYFKNEEKTKEVYKVDEKGMRWFYTGDIGRF 540
> Hs19923335
Length=671
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 67 PLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEADLV 107
PL GTS+ +T++IH P A+ Y + E E D V
Sbjct 599 PLDLGTSSPNTSQIHWTPYRAMYQYRPQNEDELELREGDRV 639
> At1g17590
Length=328
Score = 27.7 bits (60), Expect = 5.6, Method: Composition-based stats.
Identities = 10/25 (40%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 71 GTSAKSTAEIHTAPTEAISFYHHAD 95
G+S A+IH++P++A +H+AD
Sbjct 113 GSSTAGIADIHSSPSKANFSFHYAD 137
> Hs22043654
Length=239
Score = 27.3 bits (59), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 17/61 (27%), Positives = 28/61 (45%), Gaps = 2/61 (3%)
Query 45 GRLTADMPSGAGVCVLSLNGDLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEA 104
G +T D+ +G G+ + N D L G K+ I + E H++ G +L EE
Sbjct 106 GDVTGDLRNGVGLLTPTSNSDFSL--GAREKALEAIGCSKIEQTQLVLHSEDGAHLSEEK 163
Query 105 D 105
+
Sbjct 164 N 164
> CE19027
Length=177
Score = 27.3 bits (59), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 26/60 (43%), Gaps = 5/60 (8%)
Query 16 ARYAPHKVWLNPGGEQAFVYGNHIVKRHVGRLTADMPSGAGVCVLSLNGDLPLGFGTSAK 75
A Y P ++L E VY + + H +T P G C L ++ D L TSAK
Sbjct 70 ADYTP--IFL---SEVPSVYTSGTLNVHFALITVSPPDELGFCTLGVDIDTTLAAATSAK 124
> 7296203
Length=1427
Score = 26.9 bits (58), Expect = 9.8, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 53 SGAGVCVLSLNGDLPLGFGTSAKSTAEIHTAPTEAIS 89
SGA + V S + L LGF +A+++A T+P + +S
Sbjct 588 SGAPIPVHSTSSSLELGFSHTAQNSALSETSPDDFLS 624
Lambda K H
0.321 0.138 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164169380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40