bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
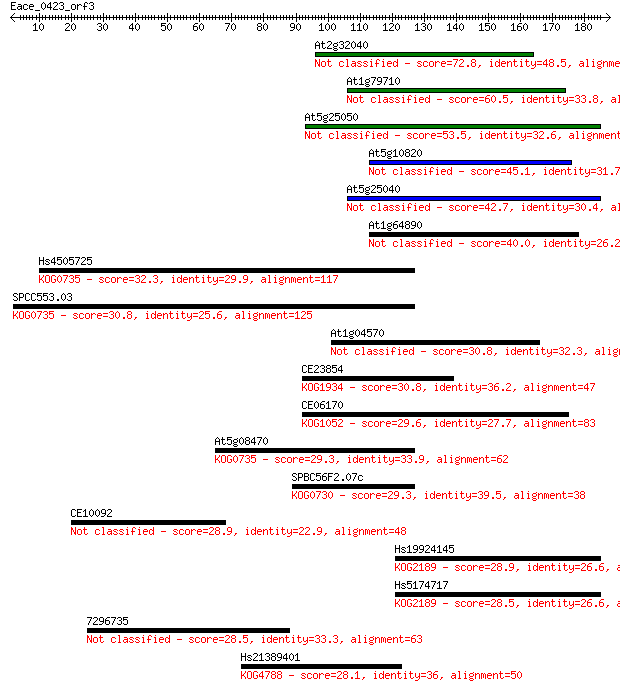
Query= Eace_0423_orf3
Length=187
Score E
Sequences producing significant alignments: (Bits) Value
At2g32040 72.8 3e-13
At1g79710 60.5 2e-09
At5g25050 53.5 2e-07
At5g10820 45.1 9e-05
At5g25040 42.7 4e-04
At1g64890 40.0 0.003
Hs4505725 32.3 0.57
SPCC553.03 30.8 1.5
At1g04570 30.8 1.5
CE23854 30.8 1.6
CE06170 29.6 3.3
At5g08470 29.3 4.3
SPBC56F2.07c 29.3 4.6
CE10092 28.9 5.6
Hs19924145 28.9 6.1
Hs5174717 28.5 7.5
7296735 28.5 8.1
Hs21389401 28.1 9.5
> At2g32040
Length=429
Score = 72.8 bits (177), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 33/68 (48%), Positives = 45/68 (66%), Gaps = 0/68 (0%)
Query 96 LIAMLQGVEVLCNLAILYLFKDDFRIDPALLAVVLGALRLPWTVKPLWALAADNFPLFGY 155
++ +QGV L LA+ + KDD +DPA AV+ G LPW VKPL+ +D+ PLFGY
Sbjct 1 MVYFVQGVLGLARLAVSFYLKDDLHLDPAETAVITGLSSLPWLVKPLYGFISDSVPLFGY 60
Query 156 RRKSYIFL 163
RR+SY+ L
Sbjct 61 RRRSYLVL 68
> At1g79710
Length=497
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 41/68 (60%), Gaps = 0/68 (0%)
Query 106 LCNLAILYLFKDDFRIDPALLAVVLGALRLPWTVKPLWALAADNFPLFGYRRKSYIFLGA 165
L ++ Y FKD+ +I P+ + +G +++PW +KPLW L D P+ GYRR+ Y L
Sbjct 67 LSKVSTQYYFKDEQKIQPSQAQIYVGLIQIPWIIKPLWGLLTDVVPVLGYRRRPYFILAG 126
Query 166 SMCIVATL 173
+ +++ +
Sbjct 127 FLAMISMM 134
> At5g25050
Length=499
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 47/93 (50%), Gaps = 1/93 (1%)
Query 93 VSSLIAMLQGVE-VLCNLAILYLFKDDFRIDPALLAVVLGALRLPWTVKPLWALAADNFP 151
V SL + QG+ L +A Y KD ++ P+ + ++PW +KPLW + D P
Sbjct 49 VVSLYGINQGLGGSLGRVATEYYMKDVQKVQPSESQALTAITKIPWIIKPLWGILTDVLP 108
Query 152 LFGYRRKSYIFLGASMCIVATLGIGLGGYKSLY 184
+FG+ R+ Y L + +V+ L I L LY
Sbjct 109 IFGFHRRPYFILAGVLGVVSLLFISLHSNLHLY 141
> At5g10820
Length=503
Score = 45.1 bits (105), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 39/63 (61%), Gaps = 0/63 (0%)
Query 113 YLFKDDFRIDPALLAVVLGALRLPWTVKPLWALAADNFPLFGYRRKSYIFLGASMCIVAT 172
Y +KD ++ P+++ + +G +PW ++P+W L D FP+ GY+RK Y + + +V+
Sbjct 82 YYWKDVQQVQPSVVQLYMGLYYIPWVMRPIWGLFTDVFPIKGYKRKPYFVVSGVLGLVSA 141
Query 173 LGI 175
+ I
Sbjct 142 IAI 144
> At5g25040
Length=492
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 40/80 (50%), Gaps = 1/80 (1%)
Query 106 LCNLAILYLFKDDFRIDPALLAVVLGALRLPWTV-KPLWALAADNFPLFGYRRKSYIFLG 164
L ++A Y KD ++ P+ + R+ W + KPL+ + D P+FG+ R+ Y L
Sbjct 63 LGHVATDYYMKDVQKVQPSQYQALSAITRISWIIFKPLFGILTDVLPIFGFHRRPYFILA 122
Query 165 ASMCIVATLGIGLGGYKSLY 184
+ +V+ L I L LY
Sbjct 123 GVIGVVSLLFISLQSNLHLY 142
> At1g64890
Length=442
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 17/65 (26%), Positives = 35/65 (53%), Gaps = 0/65 (0%)
Query 113 YLFKDDFRIDPALLAVVLGALRLPWTVKPLWALAADNFPLFGYRRKSYIFLGASMCIVAT 172
+ + R++P++L ++ + LP KP++ + +D+ FG R YI +GA + ++
Sbjct 42 FFLTEQLRVNPSVLQLLQNSANLPMVAKPIYGVVSDSVYFFGQHRIPYIAVGALLQAISW 101
Query 173 LGIGL 177
L I
Sbjct 102 LAIAF 106
> Hs4505725
Length=1283
Score = 32.3 bits (72), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 35/119 (29%), Positives = 51/119 (42%), Gaps = 25/119 (21%)
Query 10 KWGYVHWKGPHILHQVFRWVVKMEKAPGLSLPFVDAEDVSISAQHQKPTKQCS--SGEWD 67
+ ++ KGP +L +++ E+A D+ I AQ KP C E++
Sbjct 899 RMNFISVKGPELLS---KYIGASEQA---------VRDIFIRAQAAKP---CILFFDEFE 943
Query 68 SSVPEEGRALLDEKGLIPVPFATRVVSSLIAMLQGVEVLCNLAILYLFKDDFRIDPALL 126
S P G D G+ RVV+ L+ L GVE L + +L IDPALL
Sbjct 944 SIAPRRGH---DNTGV-----TDRVVNQLLTQLDGVEGLQGVYVLAATSRPDLIDPALL 994
> SPCC553.03
Length=937
Score = 30.8 bits (68), Expect = 1.5, Method: Composition-based stats.
Identities = 32/125 (25%), Positives = 53/125 (42%), Gaps = 21/125 (16%)
Query 2 SHSVHCTYKWGYVHWKGPHILHQVFRWVVKMEKAPGLSLPFVDAEDVSISAQHQKPTKQC 61
+ ++ T+ ++ KGP +L +++ K E+ D+ AQ KP
Sbjct 657 ASAISSTFPVQFISIKGPELLD---KYIGKSEQG---------VRDLFSRAQMAKPCVLF 704
Query 62 SSGEWDSSVPEEGRALLDEKGLIPVPFATRVVSSLIAMLQGVEVLCNLAILYLFKDDFRI 121
E+DS P G+ D G+ RVV+ ++ + G E L + I+ I
Sbjct 705 FD-EFDSVAPRRGQ---DSTGV-----TDRVVNQILTQMDGAESLDGVYIVAATTRPDMI 755
Query 122 DPALL 126
DPALL
Sbjct 756 DPALL 760
> At1g04570
Length=492
Score = 30.8 bits (68), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 21/78 (26%), Positives = 34/78 (43%), Gaps = 13/78 (16%)
Query 101 QGVEVLCNL-------------AILYLFKDDFRIDPALLAVVLGALRLPWTVKPLWALAA 147
+G+ VLC L A+ + + P+ L +V + LP KPL+ + +
Sbjct 41 KGISVLCGLGYWVQGSRCFPWLALNFHMVHSLALQPSTLQLVQYSCSLPMVAKPLYGVLS 100
Query 148 DNFPLFGYRRKSYIFLGA 165
D + RR YI +GA
Sbjct 101 DVLYIGSGRRVPYIAIGA 118
> CE23854
Length=1110
Score = 30.8 bits (68), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 92 VVSSLIAMLQGVEVLCNLAILYLFKDDFRIDPALLAVVLGALRLPWT 138
V +S+IA + VE + A L + +FR +P +A +LG L L T
Sbjct 948 VATSIIASIMTVESVLQRAYLLMVHQNFRPNPNFVAGILGILSLTGT 994
> CE06170
Length=956
Score = 29.6 bits (65), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 23/84 (27%), Positives = 41/84 (48%), Gaps = 6/84 (7%)
Query 92 VVSSLIAMLQGVEVLC-NLAILYLFKDDFRIDPALLAVVLGALRLPWTVKPLWALAADNF 150
+ +LIA L ++ C +AI D+ ++P L V +P+ KPLW + ++F
Sbjct 4 IAKTLIAFLILLKTDCYKIAIPANLIDE--VNPVLEFVDFRVQVIPYETKPLWRIKQESF 61
Query 151 PLFGYRRKSYIFLGASMCIVATLG 174
+ G ++ +F S+C LG
Sbjct 62 KIVGISIENGLF---SVCNCLILG 82
> At5g08470
Length=1125
Score = 29.3 bits (64), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 29/62 (46%), Gaps = 8/62 (12%)
Query 65 EWDSSVPEEGRALLDEKGLIPVPFATRVVSSLIAMLQGVEVLCNLAILYLFKDDFRIDPA 124
E+DS P+ G D G+ RVV+ + L GVEVL + + +DPA
Sbjct 945 EFDSIAPKRGH---DNTGV-----TDRVVNQFLTELDGVEVLTGVFVFAATSRPDLLDPA 996
Query 125 LL 126
LL
Sbjct 997 LL 998
> SPBC56F2.07c
Length=809
Score = 29.3 bits (64), Expect = 4.6, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 89 ATRVVSSLIAMLQGVEVLCNLAILYLFKDDFRIDPALL 126
+ RVV++L+ L G+E L N+ +L IDPAL+
Sbjct 663 SDRVVAALLNELDGIEALRNVLVLAATNRPDMIDPALM 700
> CE10092
Length=5105
Score = 28.9 bits (63), Expect = 5.6, Method: Composition-based stats.
Identities = 11/48 (22%), Positives = 26/48 (54%), Gaps = 0/48 (0%)
Query 20 HILHQVFRWVVKMEKAPGLSLPFVDAEDVSISAQHQKPTKQCSSGEWD 67
HI+H++++ ++ +E L + D+ + H+ +KQ G++D
Sbjct 4594 HIIHKLYKKMMYIENGREAKLAVQEFMDIGLDIDHEPFSKQVKMGQYD 4641
> Hs19924145
Length=830
Score = 28.9 bits (63), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 17/65 (26%), Positives = 33/65 (50%), Gaps = 6/65 (9%)
Query 121 IDPALLAVVLGALRLPWTVKPLWALAADNFPLFG-YRRKSYIFLGASMCIVATLGIGLGG 179
+DP + V LG P+ + P+W+LAA++ ++ K + LG + G+ LG
Sbjct 500 LDPNVTGVFLGPY--PFGIDPIWSLAANHLSFLNSFKMKMSVILG---VVHMAFGVVLGV 554
Query 180 YKSLY 184
+ ++
Sbjct 555 FNHVH 559
> Hs5174717
Length=614
Score = 28.5 bits (62), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 17/65 (26%), Positives = 33/65 (50%), Gaps = 6/65 (9%)
Query 121 IDPALLAVVLGALRLPWTVKPLWALAADNFPLFG-YRRKSYIFLGASMCIVATLGIGLGG 179
+DP + V LG P+ + P+W+LAA++ ++ K + LG + G+ LG
Sbjct 284 LDPNVTGVFLGPY--PFGIDPIWSLAANHLSFLNSFKMKMSVILGV---VHMAFGVVLGV 338
Query 180 YKSLY 184
+ ++
Sbjct 339 FNHVH 343
> 7296735
Length=1247
Score = 28.5 bits (62), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 27/63 (42%), Gaps = 4/63 (6%)
Query 25 VFRWVVKMEKAPGLSLPFVDAEDVSISAQHQKPTKQCSSGEWDSSVPEEGRALLDEKGLI 84
FR K + + GL+ P V A AQH T Q W + G + EK LI
Sbjct 1023 TFRKPEKRDMSVGLTTPRVRAH----RAQHVPKTLQEGQKSWRPWKQQSGEKSVTEKLLI 1078
Query 85 PVP 87
P+P
Sbjct 1079 PLP 1081
> Hs21389401
Length=182
Score = 28.1 bits (61), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 26/50 (52%), Gaps = 0/50 (0%)
Query 73 EGRALLDEKGLIPVPFATRVVSSLIAMLQGVEVLCNLAILYLFKDDFRID 122
+GR LL E GL + F V SS A L + LA+ +LF D +++
Sbjct 41 KGRLLLAESGLSFITFICYVASSASAFLTAPLLEFLLALYFLFADAMQLN 90
Lambda K H
0.324 0.139 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3058524910
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40