bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
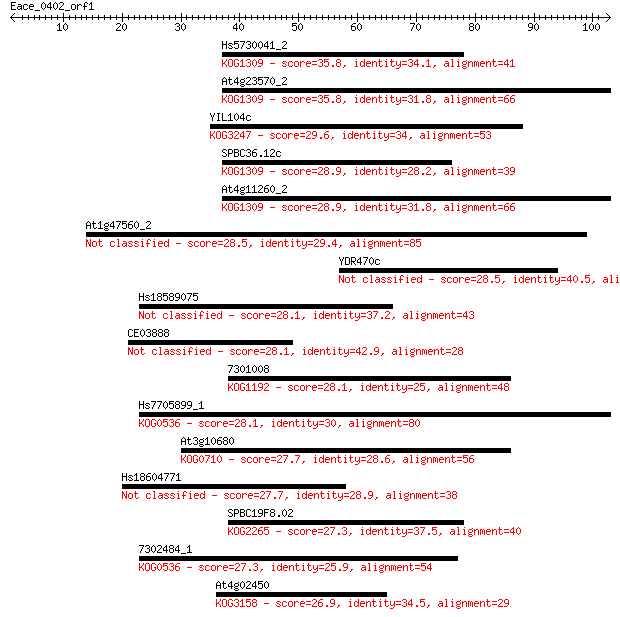
Query= Eace_0402_orf1
Length=102
Score E
Sequences producing significant alignments: (Bits) Value
Hs5730041_2 35.8 0.019
At4g23570_2 35.8 0.022
YIL104c 29.6 1.3
SPBC36.12c 28.9 2.4
At4g11260_2 28.9 2.5
At1g47560_2 28.5 3.0
YDR470c 28.5 3.3
Hs18589075 28.1 3.6
CE03888 28.1 3.7
7301008 28.1 4.0
Hs7705899_1 28.1 4.5
At3g10680 27.7 4.9
Hs18604771 27.7 5.5
SPBC19F8.02 27.3 6.3
7302484_1 27.3 6.3
At4g02450 26.9 8.9
> Hs5730041_2
Length=196
Score = 35.8 bits (81), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 37 RYEWMQNAEKIFITFFVKSLQPSDVNVDISPQSLKVQIQNP 77
+Y+W Q ++ IT +K++Q +DVNV+ S + L ++ P
Sbjct 4 KYDWYQTESQVVITLMIKNVQKNDVNVEFSEKELSALVKLP 44
> At4g23570_2
Length=202
Score = 35.8 bits (81), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 37/67 (55%), Gaps = 3/67 (4%)
Query 37 RYEWMQNAEKIFITFFVKSLQPSDVNVDISPQSLKVQIQNPLINQEAAKNQQQLYEFHV- 95
R+E+ Q E++ +T F K + +VN+D Q L V I+ P ++A Q +L+ +
Sbjct 5 RHEYYQKPEEVVVTVFAKGIPKQNVNIDFGEQILSVVIEVP--GEDAYYLQPRLFGKIIP 62
Query 96 QKLRHEV 102
K ++EV
Sbjct 63 DKCKYEV 69
> YIL104c
Length=507
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 33/64 (51%), Gaps = 11/64 (17%)
Query 35 TPRYEWMQNAEKIFITFFVKSLQPSDVNVDI-----------SPQSLKVQIQNPLINQEA 83
TPR+ Q+ E IF+ F+ +++ S V ++I SP L+++ + LI+ E
Sbjct 3 TPRFSITQDEEFIFLKIFISNIRFSAVGLEIIIQENMIIFHLSPYYLRLRFPHELIDDER 62
Query 84 AKNQ 87
+ Q
Sbjct 63 STAQ 66
> SPBC36.12c
Length=379
Score = 28.9 bits (63), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 11/39 (28%), Positives = 24/39 (61%), Gaps = 0/39 (0%)
Query 37 RYEWMQNAEKIFITFFVKSLQPSDVNVDISPQSLKVQIQ 75
RY+W Q + + I + K ++ DV++ + +LK++I+
Sbjct 185 RYDWSQTSFSLNIDIYAKKVKDEDVSLLMEKNTLKIEIK 223
> At4g11260_2
Length=198
Score = 28.9 bits (63), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 34/67 (50%), Gaps = 3/67 (4%)
Query 37 RYEWMQNAEKIFITFFVKSLQPSDVNVDISPQSLKVQIQNPLINQEAAKNQQQLYEFHV- 95
R+E+ Q E+ +T F K + +V V+ Q L V I + +EA Q +L+ +
Sbjct 1 RHEFYQKPEEAVVTIFAKKVPKENVTVEFGEQILSVVID--VAGEEAYHLQPRLFGKIIP 58
Query 96 QKLRHEV 102
+K R EV
Sbjct 59 EKCRFEV 65
> At1g47560_2
Length=672
Score = 28.5 bits (62), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 25/87 (28%), Positives = 40/87 (45%), Gaps = 16/87 (18%)
Query 14 NSSSSATTMQTTSETASASSFTPRYEWMQNAEKIFITFFVKSLQPSDVNVDISPQSLKVQ 73
NS+ SA T + + S R +W+ E+ F ++D +P+ LKV
Sbjct 64 NSNYSAVTRSSKLDFHRFSGDRVR-DWLFKLEQFF-------------SLDFTPEELKVS 109
Query 74 IQNPLINQEAAKNQQQLYE--FHVQKL 98
I + L + AAK + L+E F V+ L
Sbjct 110 IASTLCDGAAAKWYKSLFESDFGVKLL 136
> YDR470c
Length=502
Score = 28.5 bits (62), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 25/44 (56%), Gaps = 7/44 (15%)
Query 57 QPSDVNVDISPQSLK-VQIQNPLINQEA------AKNQQQLYEF 93
+P+D+++ I+PQSL + + N L +QE A N +Y F
Sbjct 214 EPTDISLTIAPQSLHTIDVINALFDQEGIRGLWKANNTTFIYNF 257
> Hs18589075
Length=247
Score = 28.1 bits (61), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 27/49 (55%), Gaps = 6/49 (12%)
Query 23 QTTSETASASSFT-PRYEWMQNAEKIFIT-----FFVKSLQPSDVNVDI 65
Q + T +A+S T PR ++Q+ E +F++ +F SL D NV +
Sbjct 77 QVSMPTTAATSITSPRVLFLQDLEALFVSLTTGKYFKMSLHDDDANVTV 125
> CE03888
Length=244
Score = 28.1 bits (61), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 21 TMQTTSETASASSFTPRYEWMQNAEKIF 48
+ ++ A+ S+TPR E +Q AEKIF
Sbjct 138 VLDNRNQAANVLSYTPRSEALQKAEKIF 165
> 7301008
Length=540
Score = 28.1 bits (61), Expect = 4.0, Method: Composition-based stats.
Identities = 12/48 (25%), Positives = 25/48 (52%), Gaps = 0/48 (0%)
Query 38 YEWMQNAEKIFITFFVKSLQPSDVNVDISPQSLKVQIQNPLINQEAAK 85
++ N ++ + K L P+D+N D +++K ++NP Q A +
Sbjct 390 FDQFNNIHRVQLAGMAKVLDPNDLNADTLIETIKELLENPSYAQRAKE 437
> Hs7705899_1
Length=244
Score = 28.1 bits (61), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 32/80 (40%), Gaps = 9/80 (11%)
Query 23 QTTSETASASSFTPRYEWMQNAEKIFITFFVKSLQPSDVNVDISPQSLKVQIQNPLINQE 82
Q T A P Y+W Q + I + K D+N+D S+ V QN E
Sbjct 121 QVTDTLAKEGPSYPSYDWFQTDSLVTIAIYTKQ---KDINLD----SIIVDHQNDSFRAE 173
Query 83 AAKNQQQLYEFHVQKLRHEV 102
+ LY H+ L HEV
Sbjct 174 TII-KDCLYLIHIG-LSHEV 191
> At3g10680
Length=490
Score = 27.7 bits (60), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 16/61 (26%), Positives = 30/61 (49%), Gaps = 8/61 (13%)
Query 30 SASSFTPRYEWMQNAEKIFI-----TFFVKSLQPSDVNVDISPQSLKVQIQNPLINQEAA 84
+ SSF PR +W + IF+ F+ ++ + D +++++Q Q PL Q A
Sbjct 22 TVSSFKPRAQWTNSGSSIFLYVNLPGFYRDQIE---IKKDERTRTVQIQGQRPLSAQTKA 78
Query 85 K 85
+
Sbjct 79 R 79
> Hs18604771
Length=409
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 11/38 (28%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 20 TTMQTTSETASASSFTPRYEWMQNAEKIFITFFVKSLQ 57
+ ++ +SE A++ S T + W+QN ++FI S++
Sbjct 102 SALKPSSEGATSVSLTVSHTWLQNRTEVFIESVCPSVE 139
> SPBC19F8.02
Length=166
Score = 27.3 bits (59), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 20/41 (48%), Gaps = 1/41 (2%)
Query 38 YEWMQNAEKIFITFFV-KSLQPSDVNVDISPQSLKVQIQNP 77
YEW Q + I V K + + VD+S LK+QI P
Sbjct 11 YEWDQTIADVDIVIHVPKGTRAKSLQVDMSNHDLKIQINVP 51
> 7302484_1
Length=287
Score = 27.3 bits (59), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 14/57 (24%), Positives = 29/57 (50%), Gaps = 3/57 (5%)
Query 23 QTTSETASASSFTPRYEWMQNAEKIFITFFVKSLQPSDVNV---DISPQSLKVQIQN 76
Q T+ PR++W+Q ++ + F+ +SL + V D+ S++V +Q+
Sbjct 175 QITNVILQPPEIVPRFDWIQQRSELTLIFYTRSLANPGLVVRRKDLQQLSVRVLVQH 231
> At4g02450
Length=262
Score = 26.9 bits (58), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 10/29 (34%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 36 PRYEWMQNAEKIFITFFVKSLQPSDVNVD 64
P +W + EKIF+T + + + VN+D
Sbjct 9 PEVKWAETTEKIFLTVVLADTKDTKVNLD 37
Lambda K H
0.310 0.121 0.324
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1181107380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40