bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0401_orf2
Length=233
Score E
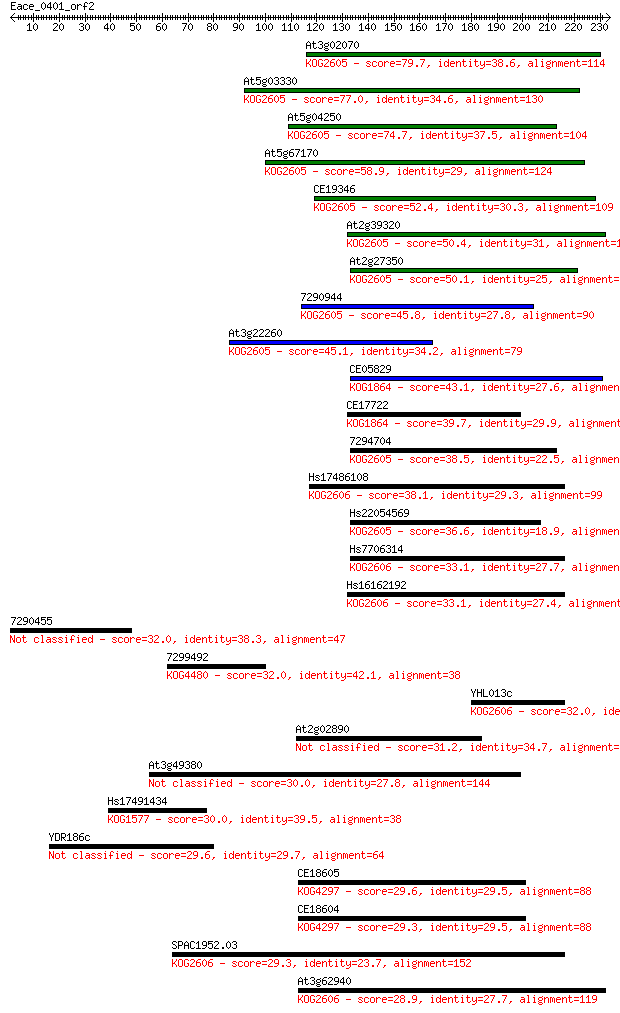
Sequences producing significant alignments: (Bits) Value
At3g02070 79.7 4e-15
At5g03330 77.0 3e-14
At5g04250 74.7 1e-13
At5g67170 58.9 8e-09
CE19346 52.4 8e-07
At2g39320 50.4 3e-06
At2g27350 50.1 4e-06
7290944 45.8 8e-05
At3g22260 45.1 1e-04
CE05829 43.1 5e-04
CE17722 39.7 0.005
7294704 38.5 0.010
Hs17486108 38.1 0.016
Hs22054569 36.6 0.043
Hs7706314 33.1 0.43
Hs16162192 33.1 0.48
7290455 32.0 1.1
7299492 32.0 1.1
YHL013c 32.0 1.1
At2g02890 31.2 2.0
At3g49380 30.0 4.7
Hs17491434 30.0 4.8
YDR186c 29.6 5.2
CE18605 29.6 5.2
CE18604 29.3 7.4
SPAC1952.03 29.3 8.0
At3g62940 28.9 8.2
> At3g02070
Length=219
Score = 79.7 bits (195), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 44/118 (37%), Positives = 64/118 (54%), Gaps = 6/118 (5%)
Query 116 LEQRLSFLGCRSVTSVGDGNCQFRSCALNLFGSERFHMLVRREAVAQMQRQREVYAQFFE 175
L QRL+ G + GDGNCQFR+ + L+ S +H VRRE V Q++ R +Y +
Sbjct 70 LLQRLNVYGLCELKVSGDGNCQFRALSDQLYRSPEYHKQVRREVVKQLKECRSMYESYV- 128
Query 176 SPQLLDRYLRDMNRKGTWGDELSLRAIADAFCCSIHLITSTPSSWY----PRYDPERG 229
P RY + M + G WGD ++L+A AD F I L+TS + + P+Y +G
Sbjct 129 -PMKYKRYYKKMGKFGEWGDHITLQAAADRFAAKICLLTSFRDTCFIEIIPQYQAPKG 185
> At5g03330
Length=356
Score = 77.0 bits (188), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 45/130 (34%), Positives = 65/130 (50%), Gaps = 4/130 (3%)
Query 92 PAAATATAEYPKSMKAWEASSNDLLEQRLSFLGCRSVTSVGDGNCQFRSCALNLFGSERF 151
P E P +A S ++ L RL V GDGNCQFR+ A L+ +
Sbjct 182 PYIPKINGEIPPEEEA--VSDHERLRNRLEMFDFTEVKVPGDGNCQFRALADQLYKTADR 239
Query 152 HMLVRREAVAQMQRQREVYAQFFESPQLLDRYLRDMNRKGTWGDELSLRAIADAFCCSIH 211
H VRR+ V Q++ + + Y + P YLR M+R G WGD ++L+A ADA+ I
Sbjct 240 HKHVRRQIVKQLKSRPDSYQGYV--PMDFSDYLRKMSRSGEWGDHVTLQAAADAYRVKIV 297
Query 212 LITSTPSSWY 221
++TS + Y
Sbjct 298 VLTSFKDTCY 307
> At5g04250
Length=395
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 39/104 (37%), Positives = 54/104 (51%), Gaps = 2/104 (1%)
Query 109 EASSNDLLEQRLSFLGCRSVTSVGDGNCQFRSCALNLFGSERFHMLVRREAVAQMQRQRE 168
+ S ++ L QRL G GDGNCQFRS + L+ S H VR + V Q+ RE
Sbjct 188 QISDHERLFQRLQLYGLVENKIEGDGNCQFRSLSDQLYRSPEHHNFVREQVVNQLAYNRE 247
Query 169 VYAQFFESPQLLDRYLRDMNRKGTWGDELSLRAIADAFCCSIHL 212
+Y + P + YL+ M R G WGD ++L+A AD + L
Sbjct 248 IYEGYV--PMAYNDYLKAMKRNGEWGDHVTLQAAADLVLTQLKL 289
> At5g67170
Length=382
Score = 58.9 bits (141), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 36/126 (28%), Positives = 57/126 (45%), Gaps = 2/126 (1%)
Query 100 EYPKSMKAWEASSNDLLEQR--LSFLGCRSVTSVGDGNCQFRSCALNLFGSERFHMLVRR 157
+ P A++ DL + R L LG + + DGNC FR+ A L G+E H R
Sbjct 17 QLPDKQLAFQGKDCDLSQFRAQLDALGLKIIQVTADGNCFFRAIADQLEGNEDEHNKYRN 76
Query 158 EAVAQMQRQREVYAQFFESPQLLDRYLRDMNRKGTWGDELSLRAIADAFCCSIHLITSTP 217
V + + RE++ F E + Y + M+ GTW + L+A + +I + +
Sbjct 77 MIVLYIVKNREMFEPFIEDDVPFEDYCKTMDDDGTWAGNMELQAASLVTRSNICIHRNMS 136
Query 218 SSWYPR 223
WY R
Sbjct 137 PRWYIR 142
> CE19346
Length=442
Score = 52.4 bits (124), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 33/117 (28%), Positives = 55/117 (47%), Gaps = 10/117 (8%)
Query 119 RLSFLGCRSVTSVGDGNCQFRSCALNLFGSERFHMLVRREAVAQMQRQREVYAQFFESPQ 178
RL+ G VGDG C FRS A ++G + H +RR + M R+ + +F +
Sbjct 130 RLAARGLIIKEMVGDGACMFRSIAEQIYGDQEMHGQIRRLCMDYMSNNRDHFKEFIT--E 187
Query 179 LLDRYLRDMNRKGTWGDELSLRAIAD-------AFCCSIHLITSTP-SSWYPRYDPE 227
+ Y++ + G+ + L+AI++ S+H I S P + PR +PE
Sbjct 188 NFENYIQRKREENVHGNHVELQAISEIKKKSNYILKNSLHSINSEPINVLLPRPEPE 244
> At2g39320
Length=189
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 31/100 (31%), Positives = 43/100 (43%), Gaps = 21/100 (21%)
Query 132 GDGNCQFRSCALNLFGSERFHMLVRREAVAQMQRQREVYAQFFESPQLLDRYLRDMNRKG 191
DGNCQFR+ A L+ + H LVR+E V Q ++
Sbjct 4 SDGNCQFRALADQLYQNSDCHELVRQEIVKQNM---------------------SLSTNS 42
Query 192 TWGDELSLRAIADAFCCSIHLITSTPSSWYPRYDPERGGE 231
WGDE++LR AD + I LITS + + P+ E
Sbjct 43 QWGDEVTLRVAADVYQVKIILITSIKLIPFMEFLPKSQKE 82
> At2g27350
Length=522
Score = 50.1 bits (118), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 22/88 (25%), Positives = 49/88 (55%), Gaps = 2/88 (2%)
Query 133 DGNCQFRSCALNLFGSERFHMLVRREAVAQMQRQREVYAQFFESPQLLDRYLRDMNRKGT 192
DGNC FR+ A ++G + L R+ + M+++R+ ++QF + YL+ R
Sbjct 270 DGNCLFRAVADQVYGDSEAYDLARQMCMDYMEQERDHFSQFI--TEGFTSYLKRKRRDKV 327
Query 193 WGDELSLRAIADAFCCSIHLITSTPSSW 220
+G+ + ++A+A+ + IH+ + + ++
Sbjct 328 YGNNVEIQALAEMYNRPIHIYSYSTGNY 355
> 7290944
Length=811
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 25/90 (27%), Positives = 44/90 (48%), Gaps = 2/90 (2%)
Query 114 DLLEQRLSFLGCRSVTSVGDGNCQFRSCALNLFGSERFHMLVRREAVAQMQRQREVYAQF 173
D +Q L G + D + FR A ++ ++ H +R E V M +R ++ +
Sbjct 18 DPYDQYLESRGLYRKHTARDASSLFRVIAEQMYDTQMLHYEIRLECVRFMTLKRRIFEK- 76
Query 174 FESPQLLDRYLRDMNRKGTWGDELSLRAIA 203
E P D Y++DM++ T+G LRA++
Sbjct 77 -EIPGDFDSYMQDMSKPKTYGTMTELRAMS 105
> At3g22260
Length=205
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 37/79 (46%), Gaps = 2/79 (2%)
Query 86 SNNSSSPAAATATAEYPKSMKAWEASSNDLLEQRLSFLGCRSVTSVGDGNCQFRSCALNL 145
S+ S P E P A ++LL RL+ G + GDGNCQFR+ A L
Sbjct 64 SHLDSIPHTPRVNREIPDINDA--TLDHELLSGRLATYGLAELQMEGDGNCQFRALADQL 121
Query 146 FGSERFHMLVRREAVAQMQ 164
F + +H VR+ V Q +
Sbjct 122 FRNADYHKHVRKHVVKQFE 140
> CE05829
Length=781
Score = 43.1 bits (100), Expect = 5e-04, Method: Composition-based stats.
Identities = 27/104 (25%), Positives = 52/104 (50%), Gaps = 9/104 (8%)
Query 133 DGNCQFRSCALNLFGSERFHMLVRREAVAQMQRQREVYAQFFESPQLLDRYLRDMNRKGT 192
DGNC +R+ + + G + HM+ R EAV + ++ E+ + + ++ ++Y+ + R G
Sbjct 563 DGNCFYRAISWWITGVQSHHMIFR-EAVGKHLKKNELMFKKYCHNEIYEKYVENAMRDGV 621
Query 193 WGDELSLRAIADAFCCSIHLITST-PSSWYPR-----YDPERGG 230
W + A+A+ ++ +IT S W P Y P +G
Sbjct 622 WATTCEIFAMANML--NVEIITYLGDSGWVPHSPQNNYPPRKGA 663
> CE17722
Length=532
Score = 39.7 bits (91), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 20/67 (29%), Positives = 37/67 (55%), Gaps = 1/67 (1%)
Query 132 GDGNCQFRSCALNLFGSERFHMLVRREAVAQMQRQREVYAQFFESPQLLDRYLRDMNRKG 191
DGNC +R+ + L G E HM+ REAV + ++ E + + ++ ++Y+ ++ R+G
Sbjct 330 ADGNCFYRAVSWWLTGVESHHMIF-REAVGKHLKKNEPKFKKYCHDEIYEKYVDNVMREG 388
Query 192 TWGDELS 198
W S
Sbjct 389 VWASTYS 395
> 7294704
Length=649
Score = 38.5 bits (88), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 18/80 (22%), Positives = 38/80 (47%), Gaps = 12/80 (15%)
Query 133 DGNCQFRSCALNLFGSERFHMLVRREAVAQMQRQREVYAQFFESPQLLDRYLRDMNRKGT 192
DG C FRS +L ++G E H RE + QF + ++ Y++ +
Sbjct 224 DGACLFRSISLQIYGDEEMH----------DHENREYFGQFV--TEDINSYIQRKRARDA 271
Query 193 WGDELSLRAIADAFCCSIHL 212
G+ + ++AI++ + ++ +
Sbjct 272 HGNHIEIQAISEIYSRTVEV 291
> Hs17486108
Length=288
Score = 38.1 bits (87), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 29/110 (26%), Positives = 49/110 (44%), Gaps = 12/110 (10%)
Query 117 EQRLSFLGCRSVTSVG---DGNCQFRSCALNLFGSERFHMLVRREAVAQMQRQREVYAQF 173
E+ + LG R + DG+C +R+ L S ML R A + M++ + + F
Sbjct 130 EKLAAILGARGLEMKAIPADGHCMYRAIQDQLVFSVSVEMLRCRTA-SYMKKHVDEFLPF 188
Query 174 FESPQLLDR--------YLRDMNRKGTWGDELSLRAIADAFCCSIHLITS 215
F +P+ D Y ++ R WG +L LRA++ I +I +
Sbjct 189 FSNPETSDSFGYDDFMIYCDNIVRTTAWGGQLELRALSHVLKTPIEVIQA 238
> Hs22054569
Length=591
Score = 36.6 bits (83), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 14/74 (18%), Positives = 37/74 (50%), Gaps = 2/74 (2%)
Query 133 DGNCQFRSCALNLFGSERFHMLVRREAVAQMQRQREVYAQFFESPQLLDRYLRDMNRKGT 192
DG C FR+ A ++G + H +VR+ + + + + ++ + + Y+ +
Sbjct 197 DGACLFRAVADQVYGDQDMHEVVRKHCMDYLMKNADYFSNYVT--EDFTTYINRKRKNNC 254
Query 193 WGDELSLRAIADAF 206
G+ + ++A+A+ +
Sbjct 255 HGNHIEMQAMAEMY 268
> Hs7706314
Length=289
Score = 33.1 bits (74), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 23/100 (23%), Positives = 39/100 (39%), Gaps = 24/100 (24%)
Query 133 DGNCQFRS---------CALNLFGSERFHMLVRREAVAQMQRQREVYAQFFESPQLLD-- 181
DG+C +++ CAL + +R + MQ E + F +P D
Sbjct 155 DGHCMYKAIEDQLKEKDCALTVVA-------LRSQTAEYMQSHVEDFLPFLTNPNTGDMY 207
Query 182 ------RYLRDMNRKGTWGDELSLRAIADAFCCSIHLITS 215
+Y D+ WG +L LRA++ I +I +
Sbjct 208 TPEEFQKYCEDIVNTAAWGGQLELRALSHILQTPIEIIQA 247
> Hs16162192
Length=293
Score = 33.1 bits (74), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 23/101 (22%), Positives = 39/101 (38%), Gaps = 24/101 (23%)
Query 132 GDGNCQFRS---------CALNLFGSERFHMLVRREAVAQMQRQREVYAQFFESPQLLD- 181
DG+C +++ CAL + +R + MQ E + F +P D
Sbjct 154 SDGHCMYKAIEDQLKEKDCALTVVA-------LRSQTAEYMQSHVEDFLPFLTNPNTGDM 206
Query 182 -------RYLRDMNRKGTWGDELSLRAIADAFCCSIHLITS 215
+Y D+ WG +L LRA++ I +I +
Sbjct 207 YTPEEFQKYCEDIVNTAAWGGQLELRALSHILQTPIEIIQA 247
> 7290455
Length=2779
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 23/48 (47%), Gaps = 1/48 (2%)
Query 1 QGHGGPHTGCPPSGGPSSGGPASAAATAPIAP-LKKCSSRVALMNVHR 47
Q GG G P S S+GGP S A P+ + K ALM +H+
Sbjct 2551 QNRGGASGGNPASTTVSAGGPPSLTANEPLPEYVLKADLDYALMMLHQ 2598
> 7299492
Length=296
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Query 62 QEKQKPRLI-RRLWLFRKNSSEDQPSNNSSSPAAATATA 99
Q QK R+I R++W+F KN E+Q ATA A
Sbjct 149 QMLQKKRMIARKMWIFSKNDDEEQQKQADKEAELATARA 187
> YHL013c
Length=307
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 11/36 (30%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 180 LDRYLRDMNRKGTWGDELSLRAIADAFCCSIHLITS 215
+D Y ++M WG E+ + A++ F C I ++ S
Sbjct 235 IDEYTKEMEHTAQWGGEIEILALSHVFDCPISILMS 270
> At2g02890
Length=531
Score = 31.2 bits (69), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 38/81 (46%), Gaps = 13/81 (16%)
Query 112 SNDLLEQRLSFLGCRSVTSVGDGNCQFRSCALNLFGSERFHMLVRRE---------AVAQ 162
+NDL+E+ LS L +SV C + CA ++F S F L + A+A
Sbjct 147 TNDLIEEILSRLHSKSVARF---RCVSKQCA-SMFASPYFKKLFQTRSSAKPSLLFAIAD 202
Query 163 MQRQREVYAQFFESPQLLDRY 183
+ + +FF SPQL + Y
Sbjct 203 YGIEEDYSMKFFSSPQLENTY 223
> At3g49380
Length=352
Score = 30.0 bits (66), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 40/154 (25%), Positives = 66/154 (42%), Gaps = 15/154 (9%)
Query 55 LHEMGEEQEKQKPRLIRRLWLFRKNSSE----DQPSNNSSSPAAATATAEYPKSMKAWEA 110
L E E++ Q+P+ +R WLF+K SS+ + N +S+ A A E K++ A
Sbjct 36 LVEEVEDELHQRPKRRKRRWLFKKVSSDPCAINVGINTTSTAINAIAAEETEKTVSP--A 93
Query 111 SSNDLLEQRLSFLGCRSVTSV----GDGNCQFRSCALNLFGSERFHMLVRREAVAQMQRQ 166
+ + R S R V ++ C R+ L G + LVR V +R+
Sbjct 94 AKETVFFCRTSVYLKRHVAAILIQTAFRGCLARTAVRALKGVVKLQALVRGHNV---RRR 150
Query 167 REVYAQFFESPQLLDRYLRDMNRKGT--WGDELS 198
+ Q ++ + D +K T GDE+S
Sbjct 151 TSITLQRVQALVRIQALALDHRKKLTTKLGDEIS 184
> Hs17491434
Length=117
Score = 30.0 bits (66), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 26/38 (68%), Gaps = 2/38 (5%)
Query 39 RVALMNVHRHRSDSLGLHEMGEEQEKQKPRLIRRLWLF 76
+VA+ V+RH D +H+ ++QE+ K +++RR WLF
Sbjct 28 KVAINTVYRH-IDCSHVHQ-NKDQEQLKEQVVRREWLF 63
> YDR186c
Length=877
Score = 29.6 bits (65), Expect = 5.2, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 30/64 (46%), Gaps = 4/64 (6%)
Query 16 PSSGGPASAAATAPIAPLKKCSSRVALMNVHRHRSDSLGLHEMGEEQEKQKPRLIRRLWL 75
P+S P SA A++P K+ +S NV+ H D G + ++ LI+R L
Sbjct 557 PNSQQPDSAGASSP----KRSTSSNHFTNVYVHDGDFDGTDTINNKKNLSSATLIKRKSL 612
Query 76 FRKN 79
+N
Sbjct 613 MNRN 616
> CE18605
Length=676
Score = 29.6 bits (65), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 26/91 (28%), Positives = 37/91 (40%), Gaps = 9/91 (9%)
Query 113 NDLLEQRLSFLGCRSVTSVGD---GNCQFRSCALNLFGSERFHMLVRREAVAQMQRQREV 169
ND L+Q L + RS +++GD F +C E LV R A REV
Sbjct 326 NDFLDQLLEAIHSRSPSAIGDLIYDKYIFEACDRTFSKEEIIQRLVNRPA------DREV 379
Query 170 YAQFFESPQLLDRYLRDMNRKGTWGDELSLR 200
E +L + YL D+ G + +R
Sbjct 380 EFDVIEVIELPEYYLVDITVTGLRSVPIDIR 410
> CE18604
Length=368
Score = 29.3 bits (64), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 26/91 (28%), Positives = 37/91 (40%), Gaps = 9/91 (9%)
Query 113 NDLLEQRLSFLGCRSVTSVGD---GNCQFRSCALNLFGSERFHMLVRREAVAQMQRQREV 169
ND L+Q L + RS +++GD F +C E LV R A REV
Sbjct 208 NDFLDQLLEAIHSRSPSAIGDLIYDKYIFEACDRTFSKEEIIQRLVNRPA------DREV 261
Query 170 YAQFFESPQLLDRYLRDMNRKGTWGDELSLR 200
E +L + YL D+ G + +R
Sbjct 262 EFDVIEVIELPEYYLVDITVTGLRSVPIDIR 292
> SPAC1952.03
Length=324
Score = 29.3 bits (64), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 36/160 (22%), Positives = 66/160 (41%), Gaps = 23/160 (14%)
Query 64 KQKPRLIRRLWLFRKNSSEDQPSNNSSSPAAATATAEYPKSMKAWEASSNDLLEQRLSFL 123
+QK RL RR +K S + + S A E K K E +
Sbjct 139 RQKERLERRKAEMKKMSEQAELE---SEKMADLKNEEKKKFSKILEEA------------ 183
Query 124 GCRSVTSVGDGNCQFRSCA--LNLFGSERFH-MLVRREAVAQMQRQREVYAQFF---ESP 177
G +V DGNC F S + LN + + + +R ++ + + E + F ES
Sbjct 184 GLVAVDIPADGNCLFASISHQLNYHHNVKLNSQALRNKSADYVLKHCEQFEGFLLDEESG 243
Query 178 QLL--DRYLRDMNRKGTWGDELSLRAIADAFCCSIHLITS 215
++L Y ++ WG ++ ++A+A++ +H+ +
Sbjct 244 EVLPVSDYCNEIRNNSKWGSDIEIQALANSLEVPVHVYNT 283
> At3g62940
Length=332
Score = 28.9 bits (63), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 33/135 (24%), Positives = 53/135 (39%), Gaps = 22/135 (16%)
Query 113 NDLLEQRLSFLGCRSVTSVGDGNCQFRSCALNLF-----GSERFHMLVRREAVAQMQRQR 167
N LE++L LG DG+C +R+ L S + +R A + M+ +
Sbjct 169 NAKLEKKLKPLGLTVSEIKPDGHCLYRAVENQLANRSGGASPYTYQNLREMAASYMREHK 228
Query 168 EVYAQFFESP-----------QLLDRYLRDMNRKGTWGDELSLRAIADAFCCSIHLITST 216
+ FF S + ++Y R++ WG +L L A+ C H+
Sbjct 229 TDFLPFFLSETEGDSNSGSAEERFEKYCREVESTAAWGSQLELGAL--THCLRKHI--KV 284
Query 217 PSSWYPRYDPERGGE 231
S +P D E G E
Sbjct 285 YSGSFP--DVEMGKE 297
Lambda K H
0.316 0.130 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4562417408
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40