bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
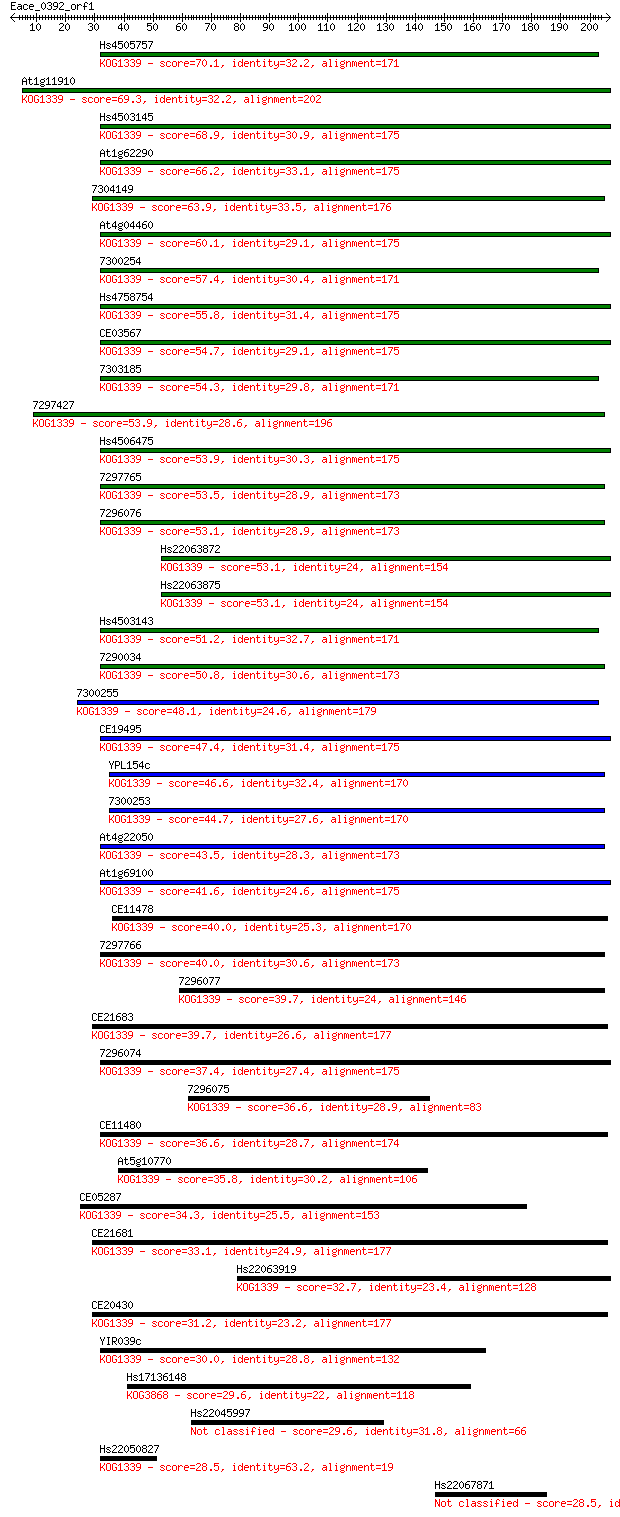
Query= Eace_0392_orf1
Length=206
Score E
Sequences producing significant alignments: (Bits) Value
Hs4505757 70.1 3e-12
At1g11910 69.3 5e-12
Hs4503145 68.9 7e-12
At1g62290 66.2 5e-11
7304149 63.9 2e-10
At4g04460 60.1 3e-09
7300254 57.4 2e-08
Hs4758754 55.8 5e-08
CE03567 54.7 1e-07
7303185 54.3 2e-07
7297427 53.9 2e-07
Hs4506475 53.9 2e-07
7297765 53.5 3e-07
7296076 53.1 3e-07
Hs22063872 53.1 4e-07
Hs22063875 53.1 4e-07
Hs4503143 51.2 2e-06
7290034 50.8 2e-06
7300255 48.1 1e-05
CE19495 47.4 2e-05
YPL154c 46.6 4e-05
7300253 44.7 1e-04
At4g22050 43.5 3e-04
At1g69100 41.6 0.001
CE11478 40.0 0.003
7297766 40.0 0.003
7296077 39.7 0.004
CE21683 39.7 0.004
7296074 37.4 0.019
7296075 36.6 0.033
CE11480 36.6 0.034
At5g10770 35.8 0.055
CE05287 34.3 0.18
CE21681 33.1 0.41
Hs22063919 32.7 0.53
CE20430 31.2 1.7
YIR039c 30.0 3.3
Hs17136148 29.6 4.3
Hs22045997 29.6 5.0
Hs22050827 28.5 8.7
Hs22067871 28.5 9.3
> Hs4505757
Length=388
Score = 70.1 bits (170), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 55/198 (27%), Positives = 85/198 (42%), Gaps = 34/198 (17%)
Query 32 FAAGSSNTWVPSISCHVTSC-------------------------ASGYIESVTGRDSLH 66
F GSSN WVPS+ C +C SG + G D+L
Sbjct 90 FDTGSSNLWVPSVYCQSQACTSHSRFNPSESSTYSTNGQTFSLQYGSGSLTGFFGYDTLT 149
Query 67 LENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQHE-QGSSSLIHLLLEAGAIKRAM 125
+++ + + FG +P F FDGI GL P +++ + +++ GA+ +
Sbjct 150 VQSIQVPNQEFGLSENEPGTNFVYAQFDGIMGLAYPALSVDEATTAMQGMVQEGALTSPV 209
Query 126 FSIALP-LSASSAGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLIDEATV 184
FS+ L SS G + FGG + S T I W P+ W++ + + LI
Sbjct 210 FSVYLSNQQGSSGGAVVFGGVDSSLY---TGQIYWAPV--TQELYWQIGIEEFLIGGQAS 264
Query 185 GVCTPQLPCTAVIDTGTS 202
G C+ C A++DTGTS
Sbjct 265 GWCSE--GCQAIVDTGTS 280
> At1g11910
Length=506
Score = 69.3 bits (168), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 65/251 (25%), Positives = 102/251 (40%), Gaps = 56/251 (22%)
Query 5 NHSESSLTRVRVGALSAVYVAVLKRYY--------------------FAAGSSNTWVPSI 44
+ E L R+G V VLK Y F GSSN WVPS
Sbjct 52 SKQEKPLRAYRLGDSGDADVVVLKNYLDAQYYGEIAIGTPPQKFTVVFDTGSSNLWVPSS 111
Query 45 SCHVT-SC-------------------------ASGYIESVTGRDSLHLENATLHDFAFG 78
C+ + +C +G I D++ + + + D F
Sbjct 112 KCYFSLACLLHPKYKSSRSSTYEKNGKAAAIHYGTGAIAGFFSNDAVTVGDLVVKDQEFI 171
Query 79 FVVQQPEYPFSELPFDGIAGLGCPQHEQGSSSLI-HLLLEAGAIKRAMFSIALPLSAS-- 135
++P F FDGI GLG + G ++ + + +L+ G IK +FS L +A
Sbjct 172 EATKEPGITFVVAKFDGILGLGFQEISVGKAAPVWYNMLKQGLIKEPVFSFWLNRNADEE 231
Query 136 SAGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLIDEATVGVCTPQLPCTA 195
GE+ FGG +P++ + + P+ + W+ + DVLI A G C + C+A
Sbjct 232 EGGELVFGGVDPNHFK---GKHTYVPV--TQKGYWQFDMGDVLIGGAPTGFC--ESGCSA 284
Query 196 VIDTGTSGIGG 206
+ D+GTS + G
Sbjct 285 IADSGTSLLAG 295
> Hs4503145
Length=396
Score = 68.9 bits (167), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 54/203 (26%), Positives = 90/203 (44%), Gaps = 36/203 (17%)
Query 32 FAAGSSNTWVPSISCHVTSC-------------------------ASGYIESVTGRDSLH 66
F GSSN WVPS+ C +C +G + + G D +
Sbjct 95 FDTGSSNLWVPSVYCTSPACKTHSRFQPSQSSTYSQPGQSFSIQYGTGSLSGIIGADQVS 154
Query 67 LENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQHEQGS-SSLIHLLLEAGAIKRAM 125
+E T+ FG V +P F + FDGI GLG P G + + ++ + M
Sbjct 155 VEGLTVVGQQFGESVTEPGQTFVDAEFDGILGLGYPSLAVGGVTPVFDNMMAQNLVDLPM 214
Query 126 FSIALPLS--ASSAGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLIDEAT 183
FS+ + + + E+ FGG++ S+ + ++ W P+ + W++ L ++ + T
Sbjct 215 FSVYMSSNPEGGAGSELIFGGYDHSHF---SGSLNWVPV--TKQAYWQIALDNIQVG-GT 268
Query 184 VGVCTPQLPCTAVIDTGTSGIGG 206
V C+ C A++DTGTS I G
Sbjct 269 VMFCSEG--CQAIVDTGTSLITG 289
> At1g62290
Length=526
Score = 66.2 bits (160), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 58/204 (28%), Positives = 88/204 (43%), Gaps = 36/204 (17%)
Query 32 FAAGSSNTWVPSISCHVT-SC-------------------------ASGYIESVTGRDSL 65
F GSSN WVPS C + SC SG I D++
Sbjct 106 FDTGSSNLWVPSGKCFFSLSCYFHAKYKSSRSSTYKKSGKRAAIHYGSGSISGFFSYDAV 165
Query 66 HLENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQHEQGSSSLI-HLLLEAGAIKRA 124
+ + + D F +P F FDG+ GLG + G+++ + + +L+ G IKR
Sbjct 166 TVGDLVVKDQEFIETTSEPGLTFLVAKFDGLLGLGFQEIAVGNATPVWYNMLKQGLIKRP 225
Query 125 MFSIALPLSASS--AGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLIDEA 182
+FS L S GEI FGG +P + + + P+ R W+ + +VLI
Sbjct 226 VFSFWLNRDPKSEEGGEIVFGGVDPKHFR---GEHTFVPV--TQRGYWQFDMGEVLIAGE 280
Query 183 TVGVCTPQLPCTAVIDTGTSGIGG 206
+ G C C+A+ D+GTS + G
Sbjct 281 STGYCGS--GCSAIADSGTSLLAG 302
> 7304149
Length=392
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 59/209 (28%), Positives = 83/209 (39%), Gaps = 45/209 (21%)
Query 29 RYYFAAGSSNTWVPSISCHVTSCA---------------------------SGYIESVTG 61
R F GSSN WVPS CH+T+ A SG +
Sbjct 87 RVVFDTGSSNLWVPSKKCHLTNIACLMHNKYDASKSKTYTKNGTEFAIQYGSGSLSGYLS 146
Query 62 RDSLHLENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQHE-QGSSSLIHLLLEAGA 120
D++ + + D F + +P F FDGI GLG + + E G
Sbjct 147 TDTVSIAGLDIKDQTFAEALSEPGLVFVAAKFDGILGLGYNSISVDKVKPPFYAMYEQGL 206
Query 121 IKRAMFSIALPLSASS--AGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVL 178
I +FS L +S GEI FGG +P++ F LP RK + W +
Sbjct 207 ISAPVFSFYLNRDPASPEGGEIIFGGSDPNHY------TGEFTYLPVTRKAY----WQIK 256
Query 179 IDEATVG---VCTPQLPCTAVIDTGTSGI 204
+D A++G +C + C + DTGTS I
Sbjct 257 MDAASIGDLQLC--KGGCQVIADTGTSLI 283
> At4g04460
Length=508
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 51/204 (25%), Positives = 86/204 (42%), Gaps = 36/204 (17%)
Query 32 FAAGSSNTWVPSISCHVT-SC-------------------------ASGYIESVTGRDSL 65
F GSSN W+PS C+++ +C +G I D +
Sbjct 104 FDTGSSNLWIPSTKCYLSVACYFHSKYKASQSSSYRKNGKPASIRYGTGAISGYFSNDDV 163
Query 66 HLENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQHEQGSSSLI-HLLLEAGAIKRA 124
+ + + + F +P F FDGI GLG + G+S+ + + ++E G +K
Sbjct 164 KVGDIVVKEQEFIEATSEPGITFLLAKFDGILGLGFKEISVGNSTPVWYNMVEKGLVKEP 223
Query 125 MFSIALPLSASS--AGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLIDEA 182
+FS L + GEI FGG +P + + + P+ + W+ + D+ I
Sbjct 224 IFSFWLNRNPKDPEGGEIVFGGVDPKHFK---GEHTFVPV--THKGYWQFDMGDLQIAGK 278
Query 183 TVGVCTPQLPCTAVIDTGTSGIGG 206
G C C+A+ D+GTS + G
Sbjct 279 PTGYCAK--GCSAIADSGTSLLTG 300
> 7300254
Length=309
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 52/199 (26%), Positives = 86/199 (43%), Gaps = 41/199 (20%)
Query 32 FAAGSSNTWVPSISCHVTSCA---------------------------SGYIESVTGRDS 64
F GSSNTW+PS++C +++ A SG + +D+
Sbjct 20 FDTGSSNTWLPSVNCPMSNSACQNHRKYNSSRSSSYIPDGRNFTLRYGSGMVVGYLSKDT 79
Query 65 LHLENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQHEQGSSS-LIHLLLEAGAIKR 123
+H+ A L F FG + + FS + FDG+ GLG +++ + LL +++
Sbjct 80 MHIAGAELPHFTFGESLFLQHFAFSSVKFDGLVGLGLGVLSWSNTTPFLELLCAQRLLEK 139
Query 124 AMFSIALPLSASSAGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLIDEAT 183
+FS+ L EI FGGF+ S + + P +W W + I +++
Sbjct 140 CVFSVYL---RRDPREIVFGGFDESKFEGKLHYV--------PVSQWH--TWSLQISKSS 186
Query 184 VGVCTPQLPCTAVIDTGTS 202
VG A++DTGTS
Sbjct 187 VGTKQIGGKSNAILDTGTS 205
> Hs4758754
Length=420
Score = 55.8 bits (133), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 55/210 (26%), Positives = 80/210 (38%), Gaps = 48/210 (22%)
Query 32 FAAGSSNTWVPSISCHVTSC---------------------------ASGYIESVTGRDS 64
F GSSN WVPS CH S +G ++ + D
Sbjct 95 FDTGSSNLWVPSRRCHFFSVPCWLHHRFDPKASSSFQANGTKFAIQYGTGRVDGILSEDK 154
Query 65 LHLENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCP-QHEQGSSSLIHLLLEAGAIKR 123
L + FG + +P F+ FDGI GLG P +G + +L+E G + +
Sbjct 155 LTIGGIKGASVIFGEALWEPSLVFAFAHFDGILGLGFPILSVEGVRPPMDVLVEQGLLDK 214
Query 124 AMFSIALPLSAS--SAGEITFGGFNPS-YVQKGTAAIAWFPLLPNPRKKWELRLWDVLID 180
+FS L GE+ GG +P+ Y+ T P W + ++
Sbjct 215 PVFSFYLNRDPEEPDGGELVLGGSDPAHYIPPLTFVPVTVP-----------AYWQIHME 263
Query 181 EATVG----VCTPQLPCTAVIDTGTSGIGG 206
VG +C C A++DTGTS I G
Sbjct 264 RVKVGPGLTLCAKG--CAAILDTGTSLITG 291
> CE03567
Length=444
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 51/205 (24%), Positives = 79/205 (38%), Gaps = 39/205 (19%)
Query 32 FAAGSSNTWVPSISCHVTSCA---------------------------SGYIESVTGRDS 64
F GSSN W+PS C A +G ++ +DS
Sbjct 111 FDTGSSNLWIPSKKCPFYDIACMLHHRYDSKSSSTYKEDGRKMAIQYGTGSMKGFISKDS 170
Query 65 LHLENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQHE-QGSSSLIHLLLEAGAIKR 123
+ + D F +P F FDGI G+ P+ G + + L E +
Sbjct 171 VCVAGVCAEDQPFAEATSEPGITFVAAKFDGILGMAYPEIAVLGVQPVFNTLFEQKKVPS 230
Query 124 AMFSIALPLSASS--AGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLIDE 181
+FS L + S GEITFGG + ++ I + P+ + W+ ++ D ++
Sbjct 231 NLFSFWLNRNPDSEIGGEITFGGIDS---RRYVEPITYVPV--TRKGYWQFKM-DKVVGS 284
Query 182 ATVGVCTPQLPCTAVIDTGTSGIGG 206
+G C A+ DTGTS I G
Sbjct 285 GVLGCSN---GCQAIADTGTSLIAG 306
> 7303185
Length=404
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 51/200 (25%), Positives = 76/200 (38%), Gaps = 38/200 (19%)
Query 32 FAAGSSNTWVPSISCHVTSCA---------------------------SGYIESVTGRDS 64
F GSSN WVPS +C T A SG + D+
Sbjct 102 FDTGSSNLWVPSATCASTMVACRVHNRYFAKRSTSHQVRGDHFAIHYGSGSLSGFLSTDT 161
Query 65 LHLENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQ-HEQGSSSLIHLLLEAGAIKR 123
+ + + D F + P F FDGI GL Q + ++E G + +
Sbjct 162 VRVAGLEIRDQTFAEATEMPGPIFLAAKFDGIFGLAYRSISMQRIKPPFYAMMEQGLLTK 221
Query 124 AMFSIALPLSA-SSAGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLIDEA 182
+FS+ L + G I FGG NP Y + + R W++++ +I
Sbjct 222 PIFSVYLSRNGEKDGGAIFFGGSNPHYYTGNFTYVQ-----VSHRAYWQVKMDSAVI--R 274
Query 183 TVGVCTPQLPCTAVIDTGTS 202
+ +C Q C +IDTGTS
Sbjct 275 NLELC--QQGCEVIIDTGTS 292
> 7297427
Length=372
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 56/224 (25%), Positives = 96/224 (42%), Gaps = 38/224 (16%)
Query 9 SSLTRVRVGALSAVYVAVLKRYYFAAGSSNTWVPSISCHVTSC----------ASGYIE- 57
+S+ GA+S A + F +GSSN WVPS +C +C +S Y+
Sbjct 63 NSMNMAYYGAISIGTPAQSFKVLFDSGSSNLWVPSNTCKSDACLTHNQYDSSASSTYVAN 122
Query 58 -----------SVTG---RDSLHLENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQ 103
S+TG D++ + ++ F +P F++ FDGI G+
Sbjct 123 GESFSIQYGTGSLTGYLSTDTVDVNGLSIQSQTFAESTNEPGTNFNDANFDGILGMAYES 182
Query 104 HE-QGSSSLIHLLLEAGAIKRAMFSIALPLSASS--AGEITFGGFNPSYVQKGTAAIAWF 160
G + + ++ G + ++FS L +S GE+ FGG + S + A+ +
Sbjct 183 LAVDGVAPPFYNMVSQGLVDNSVFSFYLARDGTSTMGGELIFGGSDASLY---SGALTYV 239
Query 161 PLLPNPRKKWELRLWDVLIDEATVGVCTPQLPCTAVIDTGTSGI 204
P+ + + W+ + ID + +C C A+ DTGTS I
Sbjct 240 PI--SEQGYWQFTMAGSSIDGYS--LCD---DCQAIADTGTSLI 276
> Hs4506475
Length=406
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 53/206 (25%), Positives = 82/206 (39%), Gaps = 39/206 (18%)
Query 32 FAAGSSNTWVPSISCHVTSCASGYIESVTGRDS-----------LHLENATLHDF----- 75
F GSSN WVPS C A Y + DS L T+ F
Sbjct 103 FDTGSSNVWVPSSKCSRLYTACVYHKLFDASDSSSYKHNGTELTLRYSTGTVSGFLSQDI 162
Query 76 ----------AFGFVVQQPEYPFSELPFDGIAGLGCPQHEQGS-SSLIHLLLEAGAIKRA 124
FG V + P PF FDG+ G+G + G + + ++ G +K
Sbjct 163 ITVGGITVTQMFGEVTEMPALPFMLAEFDGVVGMGFIEQAIGRVTPIFDNIISQGVLKED 222
Query 125 MFSIALPL----SASSAGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLID 180
+FS S S G+I GG +P + + + L+ W++++ V +
Sbjct 223 VFSFYYNRDSENSQSLGGQIVLGGSDPQHYE---GNFHYINLIKT--GVWQIQMKGVSVG 277
Query 181 EATVGVCTPQLPCTAVIDTGTSGIGG 206
+T+ +C + C A++DTG S I G
Sbjct 278 SSTL-LC--EDGCLALVDTGASYISG 300
> 7297765
Length=423
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 50/203 (24%), Positives = 85/203 (41%), Gaps = 41/203 (20%)
Query 32 FAAGSSNTWVPSISCHVT------------SCASGYIESVTG---------------RDS 64
F GSS+ WVPS+ C T S +S ++E G D+
Sbjct 91 FDTGSSDLWVPSVKCSSTNEACQKHNKYNSSASSSHVEDGKGFSIQYGSGSLSGFLSTDT 150
Query 65 LHLENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQHEQGSSSLIHLLLEAGAIKRA 124
+ ++ + + F + +P F FDGI G+ G ++ ++ G +K
Sbjct 151 VDIDGMVIRNQTFAEAIDEPGSAFVNTIFDGIIGMAFASISGGVTTPFDNIIRQGLVKHP 210
Query 125 MFSIALPL--SASSAGEITFGGFNPSYVQKGTAAIAWFPL-LPNPRKKWELRLWDVLIDE 181
+FS+ L ++ S GE+ +GG + S + I + P+ +P W+ V I+
Sbjct 211 VFSVYLRRDGTSQSGGEVIWGGIDRSIYR---GCINYVPVSMP---AYWQFTANSVKIE- 263
Query 182 ATVGVCTPQLPCTAVIDTGTSGI 204
+ +C C A+ DTGTS I
Sbjct 264 -GILLCN---GCQAIADTGTSLI 282
> 7296076
Length=405
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 50/203 (24%), Positives = 76/203 (37%), Gaps = 40/203 (19%)
Query 32 FAAGSSNTWVPSISCHVTSCA---------------------------SGYIESVTGRDS 64
F GS+N WVPS C T A +G + D+
Sbjct 110 FDTGSANLWVPSAQCLATDVACQQHNQYNSSASSTFVSSGQNFSIQYGTGSVSGYLAIDT 169
Query 65 LHLENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQ-HEQGSSSLIHLLLEAGAIKR 123
+ + + + FG V QP F+++ FDGI G+G Q E + L E G I
Sbjct 170 VTINGLAIANQTFGEAVSQPGASFTDVAFDGILGMGYQQIAEDNVVPPFYNLYEEGLIDE 229
Query 124 AMFSIALPL--SASSAGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLIDE 181
+F L SA G++T GG + + + + + P+ + W ++
Sbjct 230 PVFGFYLARNGSAVDGGQLTLGGTDQNLI---AGEMTYTPVT-------QQGYWQFAVNN 279
Query 182 ATVGVCTPQLPCTAVIDTGTSGI 204
T C A+ DTGTS I
Sbjct 280 ITWNGTVISSGCQAIADTGTSLI 302
> Hs22063872
Length=325
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 37/155 (23%), Positives = 69/155 (44%), Gaps = 9/155 (5%)
Query 53 SGYIESVTGRDSLHLENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQ-HEQGSSSL 111
+G + + G D++ + + + FG +P PFDGI GL P G++ +
Sbjct 76 TGSMTGILGYDTVQVGGISDTNQIFGLSETEPGSFLYYAPFDGILGLAYPSISSSGATPV 135
Query 112 IHLLLEAGAIKRAMFSIALPLSASSAGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWE 171
+ G + + +FS+ L S + FGG + SY T ++ W P+ W+
Sbjct 136 FDNIWNQGLVSQDLFSVYLSADDQSGSVVIFGGIDSSYY---TGSLNWVPV--TVEGYWQ 190
Query 172 LRLWDVLIDEATVGVCTPQLPCTAVIDTGTSGIGG 206
+ + + ++ + C A++DTGTS + G
Sbjct 191 ITVDSITMNGEAIACAE---GCQAIVDTGTSLLTG 222
> Hs22063875
Length=325
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 37/155 (23%), Positives = 69/155 (44%), Gaps = 9/155 (5%)
Query 53 SGYIESVTGRDSLHLENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQ-HEQGSSSL 111
+G + + G D++ + + + FG +P PFDGI GL P G++ +
Sbjct 76 TGSMTGILGYDTVQVGGISDTNQIFGLSETEPGSFLYYAPFDGILGLAYPSISSSGATPV 135
Query 112 IHLLLEAGAIKRAMFSIALPLSASSAGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWE 171
+ G + + +FS+ L S + FGG + SY T ++ W P+ W+
Sbjct 136 FDNIWNQGLVSQDLFSVYLSADDQSGSVVIFGGIDSSYY---TGSLNWVPV--TVEGYWQ 190
Query 172 LRLWDVLIDEATVGVCTPQLPCTAVIDTGTSGIGG 206
+ + + ++ + C A++DTGTS + G
Sbjct 191 ITVDSITMNGEAIACAE---GCQAIVDTGTSLLTG 222
> Hs4503143
Length=412
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 56/215 (26%), Positives = 85/215 (39%), Gaps = 55/215 (25%)
Query 32 FAAGSSNTWVPSISCHVTSCA------------SGYIESVTGRDSLHLENATLHDF---- 75
F GSSN WVPSI C + A S Y+++ T D +H + +L +
Sbjct 96 FDTGSSNLWVPSIHCKLLDIACWIHHKYNSDKSSTYVKNGTSFD-IHYGSGSLSGYLSQD 154
Query 76 -----------------------AFGFVVQQPEYPFSELPFDGIAGLGCPQHEQGSS-SL 111
FG +QP F FDGI G+ P+ + +
Sbjct 155 TVSVPCQSASSASALGGVKVERQVFGEATKQPGITFIAAKFDGILGMAYPRISVNNVLPV 214
Query 112 IHLLLEAGAIKRAMFSIALPL--SASSAGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKK 169
L++ + + +FS L A GE+ GG + Y KG+ + L RK
Sbjct 215 FDNLMQQKLVDQNIFSFYLSRDPDAQPGGELMLGGTDSKYY-KGSLS-----YLNVTRKA 268
Query 170 WELRLWDVLIDEATV--GVCTPQLPCTAVIDTGTS 202
+ W V +D+ V G+ + C A++DTGTS
Sbjct 269 Y----WQVHLDQVEVASGLTLCKEGCEAIVDTGTS 299
> 7290034
Length=407
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 53/202 (26%), Positives = 81/202 (40%), Gaps = 39/202 (19%)
Query 32 FAAGSSNTWVPSISCHVTSC----------ASGYIE------------SVTGR---DSLH 66
F GSSN WVP SC T+C +S Y+ SV+G D +
Sbjct 94 FDTGSSNLWVPGSSCISTACQDHQVFYKNKSSTYVANGTAFSITYGTGSVSGYLSVDCVG 153
Query 67 LENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQHE-QGSSSLIHLLLEAGAIKRAM 125
T+ FG V + F + FDGI G+G P G + +++ G ++ +
Sbjct 154 FAGLTIQSQTFGEVTTEQGTNFVDAYFDGILGMGFPSLAVDGVTPTFQNMMQQGLVQSPV 213
Query 126 FSIALPLSAS---SAGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLIDEA 182
FS L + S GE+ GG +PS + ++ + ++ + W D
Sbjct 214 FSFFLRDNGSVTFYGGELILGGSDPSLY---SGSLTYVNVV-------QAAYWKFQTDYI 263
Query 183 TVGVCTPQLPCTAVIDTGTSGI 204
VG + A+ DTGTS I
Sbjct 264 KVGSTSISTFAQAIADTGTSLI 285
> 7300255
Length=395
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 44/182 (24%), Positives = 74/182 (40%), Gaps = 13/182 (7%)
Query 24 VAVLKRYYFAAGSSNTWVPSISCHVTSCASGYIESVTGRDSLHLENATLHDFAFGFVVQQ 83
VA + + A +S+T+VP + +G + +D++ + + + FG +
Sbjct 117 VACHHHHRYNASASSTFVPDGRRFSIAYGTGSLSGRLAQDTVAIGQLVVQNQTFGMATHE 176
Query 84 PEYPFSELPFDGIAGLGC-PQHEQGSSSLIHLLLEAGAIKRAMFSIALPLSAS--SAGEI 140
P F + F GI GLG P E G L + + + +FS L + S GE+
Sbjct 177 PGPTFVDTNFAGIVGLGFRPIAELGIKPLFESMCDQQLVDECVFSFYLKRNGSERKGGEL 236
Query 141 TFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLIDEATVGVCTPQLPCTAVIDTG 200
FGG + + K + ++ + PL W +D V A+ DTG
Sbjct 237 LFGGVDKT---KFSGSLTYVPLT-------HAGYWQFPLDVIEVAGTRINQNRQAIADTG 286
Query 201 TS 202
TS
Sbjct 287 TS 288
> CE19495
Length=398
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 55/211 (26%), Positives = 83/211 (39%), Gaps = 45/211 (21%)
Query 32 FAAGSSNTWVPSISC----------------HVTSC----ASGYIESVTGRDSLHLENAT 71
F GSSN WVP +C +SC AS I+ TG ++N
Sbjct 86 FDTGSSNLWVPCANCPFGDIACRMHNRFDCKKSSSCTATGASFEIQYGTGSMKGTVDNDV 145
Query 72 L---HDFAF--------GFVVQQPEYPFSELPFDGIAGLGCP--QHEQGSSSLIHLLLEA 118
+ HD + +P F FDGI G+G + S + + +
Sbjct 146 VCFGHDTTYCTDKNQGLACATSEPGITFVAAKFDGIFGMGWDTISVNKISQPMDQIFANS 205
Query 119 GAIKRAMFSIALPLSA---SSAGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLW 175
K +F+ L A ++ GEIT +P++ IAW PL+ W ++L
Sbjct 206 AICKNQLFAFWLSRDANDITNGGEITLCETDPNHY---VGNIAWEPLV--SEDYWRIKLA 260
Query 176 DVLIDEATVGVCTPQLPCTAVIDTGTSGIGG 206
V+ID G P +++DTGTS + G
Sbjct 261 SVVID----GTTYTSGPIDSIVDTGTSLLTG 287
> YPL154c
Length=405
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 55/203 (27%), Positives = 78/203 (38%), Gaps = 46/203 (22%)
Query 35 GSSNTWVPSISCHVTSC-------------------------ASGYIESVTGRDSLHLEN 69
GSSN WVPS C +C +G +E +D+L + +
Sbjct 111 GSSNLWVPSNECGSLACFLHSKYDHEASSSYKANGTEFAIQYGTGSLEGYISQDTLSIGD 170
Query 70 ATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQHEQGS------SSLIHLLLEAGAIKR 123
T+ F +P F+ FDGI GLG +++ LL+ KR
Sbjct 171 LTIPKQDFAEATSEPGLTFAFGKFDGILGLGYDTISVDKVVPPFYNAIQQDLLDE---KR 227
Query 124 AMFSIA-LPLSASSAGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLIDEA 182
F + + GE TFGG + S K I W P+ R+K W+V +
Sbjct 228 FAFYLGDTSKDTENGGEATFGGIDES---KFKGDITWLPV----RRK---AYWEVKFEGI 277
Query 183 TVGVCTPQLPCT-AVIDTGTSGI 204
+G +L A IDTGTS I
Sbjct 278 GLGDEYAELESHGAAIDTGTSLI 300
> 7300253
Length=465
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 47/198 (23%), Positives = 73/198 (36%), Gaps = 36/198 (18%)
Query 35 GSSNTWVPSISCHVTSCA-------------------------SGYIESVTGRDSLHLEN 69
GSSN WVP C +C SG + V +D++ +
Sbjct 170 GSSNIWVPGPHCKSKACKKHKQYHPAKSSTYVKNGKSFAITYGSGSVAGVLAKDTVRIAG 229
Query 70 ATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQ-HEQGSSSLIHLLLEAGAIKRAMFSI 128
+ + F ++P F FDGI GLG +L+ + I F+I
Sbjct 230 LVVTNQTFAMTTKEPGTTFVTSNFDGILGLGYRSIAVDNVKTLVQNMCSEDVITSCKFAI 289
Query 129 ALPLSASSAGE--ITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLIDEATVGV 186
+ SS+ I FG N S G+ + + P+ + W+ L D+ VG
Sbjct 290 CMKGGGSSSRGGAIIFGSSNTS-AYSGSNSYTYTPV--TKKGYWQFTLQDIY-----VGG 341
Query 187 CTPQLPCTAVIDTGTSGI 204
A++D+GTS I
Sbjct 342 TKVSGSVQAIVDSGTSLI 359
> At4g22050
Length=336
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 49/204 (24%), Positives = 88/204 (43%), Gaps = 40/204 (19%)
Query 32 FAAGSSNTWVPS----------ISCHVTSCASGYIESVT------GRDSL----HLENAT 71
F GSS+ WVPS + +++S + + E+ T G+ SL ++ T
Sbjct 63 FDTGSSSLWVPSENWLAKTENPRNRYISSASRTFKENGTKAELKYGKGSLTGFLSVDTVT 122
Query 72 L-------HDFAFGFVVQQPEYPFSELPFDGIAGLGCPQHEQGSSSLIHLLLEAGAIKRA 124
+ F G E+ F ++PFDGI GL +S+ H ++ G I +
Sbjct 123 VGGISITSQTFIEGVKTPYKEF-FKKMPFDGILGLRFTDPLNFGTSVWHSMVFQGKIAKN 181
Query 125 MFSIALPLSASS----AGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLID 180
+FSI L ++S GE+ FGG P++ + + P + + ++ +
Sbjct 182 VFSIWLRRFSNSGEINGGEVVFGGIIPAHFSGDHTYVD----VEGPGNFFAMS--NIWVG 235
Query 181 EATVGVCTPQLPCTAVIDTGTSGI 204
+C+ C A++D+G+S I
Sbjct 236 GKNTNICSSG--CKAIVDSGSSNI 257
> At1g69100
Length=343
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 43/204 (21%), Positives = 84/204 (41%), Gaps = 54/204 (26%)
Query 32 FAAGSSNTWVPSI-------------------SCHVTSCA-------SGYIESVTGRDSL 65
F GS++ WVPS +C + +G + + +D++
Sbjct 64 FDTGSTDLWVPSKEWPEETDHKHPKFDKDASKTCRLMKGGEVNIAYETGSVVGILAQDNV 123
Query 66 HLENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQHE-QGSSSLIHLLLEAGAIKRA 124
++ + F+ + P+ F + FDG+ GLG QGS + + G
Sbjct 124 NVGGVVIKSQDL-FLARNPDTYFRSVKFDGVIGLGIKSSRAQGS-----VTGDGG----- 172
Query 125 MFSIALPLSASSAGEITFGGFNPSYVQKGTAAIAWFPL-LPNPRKKWELRLWDVLID-EA 182
+ G+I FGGF+P + + P+ L + R W++++ + I+ +
Sbjct 173 --------EDPNGGQIMFGGFDPKQFK---GEHVYVPMKLSDDR--WKIKMSKIYINGKP 219
Query 183 TVGVCTPQLPCTAVIDTGTSGIGG 206
+ C + CTA++D+G++ I G
Sbjct 220 AINFCD-DVECTAMVDSGSTDIFG 242
> CE11478
Length=383
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 43/175 (24%), Positives = 76/175 (43%), Gaps = 19/175 (10%)
Query 36 SSNTWVPSISCHVTSCASGYIESVTGRDSLHLENATLHDFAFGFVVQQPEYPFSELPFDG 95
+S+T++ S V G + G+D++ + + + FG + S + FDG
Sbjct 113 ASSTFIAGTSNFVLPFNDGDVSGDLGKDNIQFAGSKIQNQDFG--IGTDATRLSGVTFDG 170
Query 96 IAGLGCPQHE-QGSSSLIHLLLEAGAIKRAMFSIALPLSA----SSAGEITFGGFNPSYV 150
+ GLG P G+S+ + LL + + +F+ S+ + GEITFG + ++
Sbjct 171 VLGLGWPATALNGTSTTMQNLLP--QLDQPLFTTYFTKSSVHNGTVGGEITFGAIDTTHC 228
Query 151 QKGTAAIAWFPLLPNPRKKWELRLWDVLIDEATVGVCTPQLPCTAVIDTGTSGIG 205
Q + I + L + LW ID + G + TA+ DT +S G
Sbjct 229 Q---SQINYVRLAYDS-------LWSYSIDGFSFGNYSRNQTDTAIPDTTSSYTG 273
> 7297766
Length=391
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 53/204 (25%), Positives = 85/204 (41%), Gaps = 42/204 (20%)
Query 32 FAAGSSNTWVPSISC------------HVTSCASGY--------IESVTGRDSLHLENA- 70
F GS+N WVPS SC + +S +S Y IE TG S L N
Sbjct 93 FDTGSANLWVPSASCPASNTACQRHNKYDSSASSTYVANGEEFAIEYGTGSLSGFLSNDI 152
Query 71 ------TLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQHE-QGSSSLIHLLLEAGAIKR 123
++ + FG + +P F + PF GI GL G + ++ G +
Sbjct 153 VTIAGISIQNQTFGEALSEPGTTFVDAPFAGILGLAFSAIAVDGVTPPFDNMISQGLLDE 212
Query 124 AMFSIALPL--SASSAGEITFGGFNPSYVQKGTAAIAWFPL-LPNPRKKWELRLWDVLID 180
+ S L +A GE+ GG + S + ++ + P+ +P W+ ++ + +
Sbjct 213 PVISFYLKRQGTAVRGGELILGGIDSSLYR---GSLTYVPVSVP---AYWQFKV-NTIKT 265
Query 181 EATVGVCTPQLPCTAVIDTGTSGI 204
T+ +C C A+ DTGTS I
Sbjct 266 NGTL-LCN---GCQAIADTGTSLI 285
> 7296077
Length=418
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 35/150 (23%), Positives = 63/150 (42%), Gaps = 14/150 (9%)
Query 59 VTGRDSLHLENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQHE-QGSSSLIHLLLE 117
+ D+ L + + + F + P FDGI GLG + G + + +LE
Sbjct 163 ILSTDTFTLGDLVIKNQTFAEINSAPTDMCKRSNFDGIIGLGFSEIALNGVETPLDNILE 222
Query 118 AGAIKRAMFSIALPLSASSA---GEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRL 174
G I +FS+ + +AS A G + GG +P+ + + + P+ ++
Sbjct 223 QGLIDEPIFSLYVNRNASDASNGGVLLLGGSDPTLY---SGCLTYVPV-------SKVGF 272
Query 175 WDVLIDEATVGVCTPQLPCTAVIDTGTSGI 204
W + + + +G C A+ D GTS I
Sbjct 273 WQITVGQVEIGSKKLCSNCQAIFDMGTSLI 302
> CE21683
Length=320
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 47/181 (25%), Positives = 74/181 (40%), Gaps = 15/181 (8%)
Query 29 RYYFAAGSSNTWVPSISCHVTSCASGYIESVTGRDSLHLENATLHDFAFGFVVQQPEYPF 88
++ F +S+T+V S SG RD++ + + + FG V + F
Sbjct 115 KHKFDTKASSTFVSETRKFSISYGSGSCSGHLARDTITMGGLKIDNQEFG-VAETLASVF 173
Query 89 SELPFDGIAGLGCP--QHEQGSSSLIHLL--LEAGAIKRAMFSIALPLSASSAGEITFGG 144
+E P DGI GLG P ++ + + +LL L+A M + + G IT+G
Sbjct 174 AEQPVDGILGLGWPALADDKVTPPMQNLLPQLDAKVFTVWMDRKLQGSNGGNGGLITYGA 233
Query 145 FNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLIDEATVGVCTPQLPCTAVIDTGTSGI 204
+ I++ PL + WE L D VG + + DTGTS I
Sbjct 234 LDSVNCADD---ISYVPL--TAKTYWEFAL-----DGFAVGSFSETKTAKVISDTGTSWI 283
Query 205 G 205
G
Sbjct 284 G 284
> 7296074
Length=393
Score = 37.4 bits (85), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 48/205 (23%), Positives = 76/205 (37%), Gaps = 44/205 (21%)
Query 32 FAAGSSNTWVPSISCH--VTSC------------------------ASGYIESVTGRDSL 65
F GSS+ WVPS C + +C SG ++ + D++
Sbjct 103 FDTGSSDFWVPSSHCRFCIKTCGNKFFRKSNSKSFRSSGTPFSITYGSGSVKGIVASDNV 162
Query 66 HLENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQHEQGSSS-LIHLLLEAGAIKRA 124
+ + + G V S FDGIAG Q S +++ +++
Sbjct 163 GFGDLKIQNQGIGLV----NISDSCSVFDGIAGFAFQQLSMTKSVPSFQQMIDQQLVEQP 218
Query 125 MFSIALPLSASSAGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLIDEATV 184
+FS L +S G + GG N S + + PL E + W +D V
Sbjct 219 IFSFHLKSGSSDGGSMILGGSNSS--------LYYGPLTYT--NVTEAKYWSFKLDFIAV 268
Query 185 ---GVCTPQLPCTAVIDTGTSGIGG 206
G + + A++DTGTS I G
Sbjct 269 HGKGSRSSRTGNKAIMDTGTSLIVG 293
> 7296075
Length=410
Score = 36.6 bits (83), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 24/87 (27%), Positives = 43/87 (49%), Gaps = 4/87 (4%)
Query 62 RDSLHLENATLHDFAFGFVVQQPEYPFSELPFDGIAGLGCPQHEQGS-SSLIHLLLEAGA 120
+D+++ ++ + F + PE F + DGI GLG S + + L+ G
Sbjct 170 QDTVNFAGYSIKNQIFAEITNAPETAFLKSQLDGILGLGFASIAINSITPPFYNLMAQGL 229
Query 121 IKRAMFSIALPLSASSA---GEITFGG 144
+ R++FSI L + ++A GE+ GG
Sbjct 230 VNRSVFSIYLNRNGTNAINGGELILGG 256
> CE11480
Length=474
Score = 36.6 bits (83), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 50/209 (23%), Positives = 80/209 (38%), Gaps = 49/209 (23%)
Query 32 FAAGSSNTWVPSISCHVTSCASG-------YIES-----VTGRDSLHLENATLH------ 73
F SSN WV + C +C G Y + V G S +L H
Sbjct 170 FDTTSSNLWVFGVECRSQNCHGGRGRRDREYNRTASSTFVAGTSSFNLPYDGGHVSGNVG 229
Query 74 ----DFAFGFVVQQPEYP--------FSELPFDGIAGLGCPQHE-QGSSSLIHLLLEAGA 120
FA GF +Q ++ F E FDG+ GLG P G+S+ + LL
Sbjct 230 KDTAQFA-GFTIQSQDFGIGTAATRLFGE-TFDGVLGLGWPATALNGTSTTMQNLLP--Q 285
Query 121 IKRAMFSIALPLS----ASSAGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWD 176
+ + +F+ S ++ G+I FG + ++ Q + + + PL N W
Sbjct 286 LDQKLFTTYFTKSNMHNGTAGGDIMFGAIDTTHCQ---SQVNYVPLAYNS-------FWS 335
Query 177 VLIDEATVGVCTPQLPCTAVIDTGTSGIG 205
+D ++G + T + DT + G
Sbjct 336 YSVDGFSIGTYSRTQTETTIPDTSSGWTG 364
> At5g10770
Length=446
Score = 35.8 bits (81), Expect = 0.055, Method: Compositional matrix adjust.
Identities = 32/117 (27%), Positives = 50/117 (42%), Gaps = 12/117 (10%)
Query 38 NTWVPSISCHVTSCASGYIESVTGRDSLHLENATLHDFAFGFVVQQPEYPFSELPFDGIA 97
N S +C S A+G S + + ++ F+ GF+ ++ + FDG+
Sbjct 157 NVSCSSAACGSLSSATGNAGSCSASNCIYGIQYGDQSFSVGFLAKEKFTLTNSDVFDGVY 216
Query 98 GLGCPQHEQGSSSLIHLLLEAGAIK-----------RAMFSIALPLSASSAGEITFG 143
GC ++ QG + + LL G K +FS LP SAS G +TFG
Sbjct 217 -FGCGENNQGLFTGVAGLLGLGRDKLSFPSQTATAYNKIFSYCLPSSASYTGHLTFG 272
> CE05287
Length=428
Score = 34.3 bits (77), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 39/162 (24%), Positives = 72/162 (44%), Gaps = 17/162 (10%)
Query 25 AVLKRYYFAAGSSNTWVPSISCHVTSCASGYIESVTGRDSLHLENATLHDFAFGFVVQQP 84
A ++ F + S+++V + + +G ++ G D+ N ++ A G V Q
Sbjct 147 ACQSKHRFNSSLSSSYVTNGQKFDMTYNTGEVKGFFGVDTFCFTNTSVC--ATGQVFGQA 204
Query 85 EY---PFSELPFDGIAGLGCPQH--EQGSSSLIHLLLEAGAIKRAMFSIAL----PLSAS 135
F++ P DGI GLG P Q + L + L+ G + + F + L P S
Sbjct 205 TTIGEAFAKQPEDGIIGLGWPALAVNQQTPPLFN-LMNQGKLDQPYFVVYLANIGPTSQI 263
Query 136 SAGEITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDV 177
+ G T GG + ++ ++ + W PL + + W+ +L V
Sbjct 264 NGGAFTVGGLDTTHC---SSNVDWVPL--STQTFWQFKLGGV 300
> CE21681
Length=396
Score = 33.1 bits (74), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 44/184 (23%), Positives = 70/184 (38%), Gaps = 19/184 (10%)
Query 29 RYYFAAGSSNTWVPSISCHVTSCASGYIESVTGRDSLHLENATLHDFAFGFVVQQPEYPF 88
+ F S T+V SG G+D L+ T+ FG + F
Sbjct 114 KQKFDTTKSTTFVKETRKFSIQYGSGSCNGYLGKDVLNFGGLTVQSQEFGVSTHLADV-F 172
Query 89 SELPFDGIAGLGCPQH--EQGSSSLIHLLLEAGAIKRAMFSIALPLSASSA-----GEIT 141
P DGI GLG P +Q + +L+ + + +F++ L + A G IT
Sbjct 173 GYQPVDGILGLGWPALAVDQVVPPMQNLIAQK-QLDAPLFTVWLDRNLQIAQGTPGGLIT 231
Query 142 FGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLIDEATVGVCTPQLPCTAVIDTGT 201
+G + K + + PL + + W+ L D VG + + DTGT
Sbjct 232 YGAIDTVNCAK---QVTYVPL--SAKTYWQFPL-----DAFAVGTYSETKKDQVISDTGT 281
Query 202 SGIG 205
S +G
Sbjct 282 SWLG 285
> Hs22063919
Length=267
Score = 32.7 bits (73), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 30/132 (22%), Positives = 52/132 (39%), Gaps = 31/132 (23%)
Query 79 FVVQQPEYPFSELPFDGIAGLGCPQHEQGSSSLIHLLLEAGAIKRAMFSIALPLSASSAG 138
V +QP + ++ + G G+G P + ++ + G+ L S
Sbjct 62 LVDEQPLENYLDMEYFGTIGIGTPAQD------FTVVFDTGSSN---------LDDQSGS 106
Query 139 EITFGGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLIDEATVG----VCTPQLPCT 194
+ FGG + SY T ++ W P+ W + +D T+ C C
Sbjct 107 VVIFGGIDSSYY---TGSLNWVPVTVE-------GYWQITVDSITMNGEAIACAEG--CQ 154
Query 195 AVIDTGTSGIGG 206
A++DTGTS + G
Sbjct 155 AIVDTGTSLLTG 166
> CE20430
Length=638
Score = 31.2 bits (69), Expect = 1.7, Method: Composition-based stats.
Identities = 41/183 (22%), Positives = 71/183 (38%), Gaps = 16/183 (8%)
Query 29 RYYFAAGSSNTWVPSISCHVTSCASGYIESVTGRDSLHLENATLHDFAFGFVVQQPEYPF 88
++ F S ++V S SG G D++ + + FG V +Y F
Sbjct 107 KHKFNTTKSTSFVKSNRKFNIQYGSGECSGYLGTDTVEIGGLKIQKQEFG-VANIVDYDF 165
Query 89 SELPFDGIAGLGCPQHEQGS-SSLIHLLLEAGAIKRAMFSIAL-----PLSASSAGEITF 142
P DGI GL P + + L+ + +F+I S G IT+
Sbjct 166 GTRPIDGIFGLAWPALSVDQVTPPMQNLVSQNQLDAPIFTIFFDKRDPDYYYPSNGLITY 225
Query 143 GGFNPSYVQKGTAAIAWFPLLPNPRKKWELRLWDVLIDEATVGVCTPQLPCTAVIDTGTS 202
GG + + A I++ P+ + + W+ ++ + V Q A++DTG+S
Sbjct 226 GGLD---TKNCNANISYVPV--SSKTYWQFKVDGFQVGTYNRTVNRDQ----AIMDTGSS 276
Query 203 GIG 205
G
Sbjct 277 WFG 279
> YIR039c
Length=537
Score = 30.0 bits (66), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 38/148 (25%), Positives = 55/148 (37%), Gaps = 25/148 (16%)
Query 32 FAAGSSNTWVPSISCHVTSCASG-YIESVTGRDSLHLENATLHDFAFGFVVQQPEYPFSE 90
F A +S+T+ + + + G Y G D + EN TL+DF FG P
Sbjct 145 FDASNSSTFENNATFFNNTYGDGTYYAGTYGTDVVSFENITLNDFTFGVSNDTIGNP--- 201
Query 91 LPFDGIAGLGCPQHE--QGSSSLIHL-------------LLEAGAIKRAMFSIALPLSAS 135
GI G+ P E G + L L G I + +S+ L +
Sbjct 202 ---SGILGISLPIAEFTDGIEYALALNRTPFIYDNFPMELKNQGKINKIAYSLFLNGPDA 258
Query 136 SAGEITFGGFNPSYVQKGTAAIAWFPLL 163
G I FG + S K T + P+L
Sbjct 259 HFGSILFGAVDKS---KYTGQLYTLPML 283
> Hs17136148
Length=470
Score = 29.6 bits (65), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 26/119 (21%), Positives = 48/119 (40%), Gaps = 15/119 (12%)
Query 41 VPSISCHVTSCASGYIESVTGRDSLHLENATLHDFAFGFVVQQPEYPFSELPFDGIAGLG 100
+P++ + S + Y++ G LH++ ATL + P LP+ +GL
Sbjct 133 LPAVDWYAVSTLTTYLQEKLGASPLHVDLATLRELKLN--ASLPALLLIRLPYTASSGLM 190
Query 101 CPQHE-QGSSSLIHLLLEAGAIKRAMFSIALPLSASSAGEITFGGFNPSYVQKGTAAIA 158
P+ G+ +I G + + S +P +A+ PS V + A +A
Sbjct 191 APREVLTGNDEVI------GQVLSTLKSEDVPYTAA------LTAVRPSRVARDVAVVA 237
> Hs22045997
Length=1074
Score = 29.6 bits (65), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 35/81 (43%), Gaps = 21/81 (25%)
Query 63 DSLHLENATLHDFAFGFVVQQPEYPFSELPFDG---------------IAGLGCPQHEQG 107
D + ++ LHD +G + QP+ P S LPF+ I G C +++
Sbjct 173 DGVQIQACPLHDGLYGLPLLQPKPPISCLPFNSHKLIAFYQPNPKRHVIEGDNCSEYQYI 232
Query 108 SSSLIHLLLEAGAIKRAMFSI 128
S+L+ G KRA ++
Sbjct 233 HSTLL------GTEKRAKVTM 247
> Hs22050827
Length=160
Score = 28.5 bits (62), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 12/19 (63%), Positives = 12/19 (63%), Gaps = 0/19 (0%)
Query 32 FAAGSSNTWVPSISCHVTS 50
F GSSN WVPS CH S
Sbjct 95 FDTGSSNLWVPSRRCHFFS 113
> Hs22067871
Length=630
Score = 28.5 bits (62), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 24/46 (52%), Gaps = 8/46 (17%)
Query 147 PSYVQKGTA----AIAWFPLL----PNPRKKWELRLWDVLIDEATV 184
PS KG+A + W P+L PNPRK + L L L+ E TV
Sbjct 567 PSRELKGSADSLPVLVWKPMLFEFTPNPRKTFSLALQVHLVSERTV 612
Lambda K H
0.319 0.135 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3694581778
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40