bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0378_orf1
Length=344
Score E
Sequences producing significant alignments: (Bits) Value
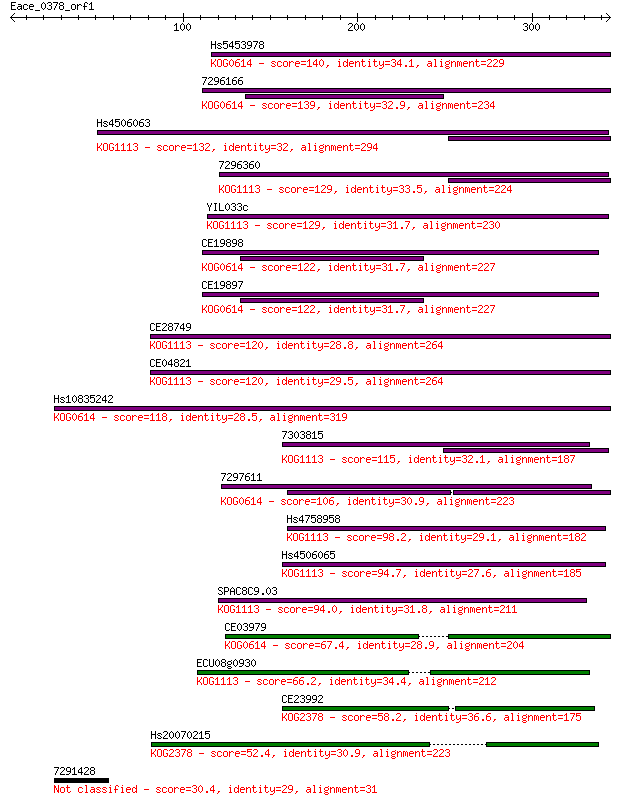
Hs5453978 140 4e-33
7296166 139 8e-33
Hs4506063 132 7e-31
7296360 129 8e-30
YIL033c 129 8e-30
CE19898 122 9e-28
CE19897 122 9e-28
CE28749 120 3e-27
CE04821 120 5e-27
Hs10835242 118 2e-26
7303815 115 1e-25
7297611 106 8e-23
Hs4758958 98.2 2e-20
Hs4506065 94.7 2e-19
SPAC8C9.03 94.0 4e-19
CE03979 67.4 4e-11
ECU08g0930 66.2 9e-11
CE23992 58.2 2e-08
Hs20070215 52.4 1e-06
7291428 30.4 5.2
> Hs5453978
Length=762
Score = 140 bits (353), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 78/236 (33%), Positives = 133/236 (56%), Gaps = 8/236 (3%)
Query 116 KTGTDLSLIRSSLSANLVCSSLNDSEVEALANAVQFFTFTKGDIVTKQGENGSYFFIVHS 175
K ++ LI +L+ N L+ +++ + + + +G + KQGE G++ F++
Sbjct 151 KDSSEKKLITDALNKNQFLKRLDPQQIKDMVECMYGRNYQQGSYIIKQGEPGNHIFVLAE 210
Query 176 GEFEVIVNDKVVNKIVEGQAFGEISLIHNSARTASIKTLSEKAALWGVQRQVFRETLKQL 235
G EV +K+++ I FGE+++++N RTAS+K ++ W + R+VF+ +++
Sbjct 211 GRLEVFQGEKLLSSIPMWTTFGELAILYNCTRTASVKAIT-NVKTWALDREVFQNIMRRT 269
Query 236 SSRNFAENRQFLASVEFFQMLTEAQKNVITNALVVQSFRAGQPIVKEGEKGDILYILKSG 295
+ + R FL SV + L E + I + L V+ + G I++EGE+G +IL G
Sbjct 270 AQARDEQYRNFLRSVSLLKNLPEDKLTKIIDCLEVEYYDKGDYIIREGEEGSTFFILAKG 329
Query 296 KARV--SINGKD----VRLLQKGDYFGERALLYDEPRSATIIAEEETV-CVSIGRD 344
K +V S G D ++ LQKG+YFGE+AL+ D+ RSA IIAEE V C+ I R+
Sbjct 330 KVKVTQSTEGHDQPQLIKTLQKGEYFGEKALISDDVRSANIIAEENDVACLVIDRE 385
> 7296166
Length=768
Score = 139 bits (351), Expect = 8e-33, Method: Compositional matrix adjust.
Identities = 77/242 (31%), Positives = 139/242 (57%), Gaps = 8/242 (3%)
Query 111 LEKREKTGTDLSLIRSSLSANLVCSSLNDSEVEALANAVQFFTFTKGDIVTKQGENGSYF 170
+ K EK +D I+ ++ N +++ S+V L +++ + G+ V ++GE G++
Sbjct 163 IPKYEKDFSDKQQIKDAIMDNDFLKNIDASQVRELVDSMYSKSIAAGEFVIREGEVGAHL 222
Query 171 FIVHSGEFEVIVNDKVVNKIVEGQAFGEISLIHNSARTASIKTLSEKAALWGVQRQVFRE 230
++ +GEF V+ KV++K+ G+AFGE+++++N RTASI+ LSE A +W + R+VF++
Sbjct 223 YVSAAGEFAVMQQGKVLDKMGAGKAFGELAILYNCTRTASIRVLSEAARVWVLDRRVFQQ 282
Query 231 TLKQLSSRNFAENRQFLASVEFFQMLTEAQKNVITNALVVQSFRAGQPIVKEGEKGDILY 290
+ + + FL SV L+E I + L ++ + AG I+++G GD +
Sbjct 283 IMMCTGLQRIENSVNFLRSVPLLMNLSEELLAKIADVLELEFYAAGTYIIRQGTAGDSFF 342
Query 291 ILKSGKARV-------SINGKDVRLLQKGDYFGERALLYDEPRSATIIAEEETV-CVSIG 342
++ G RV S ++R L +GDYFGE+AL+ ++ R+A IIA V C+++
Sbjct 343 LISQGNVRVTQKLTPTSPEETELRTLSRGDYFGEQALINEDKRTANIIALSPGVECLTLD 402
Query 343 RD 344
RD
Sbjct 403 RD 404
Score = 39.7 bits (91), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 28/123 (22%), Positives = 55/123 (44%), Gaps = 10/123 (8%)
Query 136 SLNDSEVEALANAVQFFTFTKGDIVTKQGENGSYFFIVHSGEFEVI-------VNDKVVN 188
+L++ + +A+ ++ + G + +QG G FF++ G V + +
Sbjct 307 NLSEELLAKIADVLELEFYAAGTYIIRQGTAGDSFFLISQGNVRVTQKLTPTSPEETELR 366
Query 189 KIVEGQAFGEISLIHNSARTASIKTLSEKAALWGVQRQVFRET---LKQLSSRNFAENRQ 245
+ G FGE +LI+ RTA+I LS + R F+ L +L +++ + +
Sbjct 367 TLSRGDYFGEQALINEDKRTANIIALSPGVECLTLDRDSFKRLIGDLCELKEKDYGDESR 426
Query 246 FLA 248
LA
Sbjct 427 KLA 429
> Hs4506063
Length=381
Score = 132 bits (333), Expect = 7e-31, Method: Compositional matrix adjust.
Identities = 93/304 (30%), Positives = 160/304 (52%), Gaps = 20/304 (6%)
Query 51 QQQEPQQQQEQQQPPDRKTSQPQQSDDAAAPPKPG----GERKVQKAIMQQDDTQAEDAR 106
+++E +Q Q Q+ R S+ +D +PP P G R+ + AI + T+ + A
Sbjct 58 EKEEAKQIQNLQKAGTRTDSR----EDEISPPPPNPVVKGRRR-RGAISAEVYTEEDAAS 112
Query 107 LLSH-LEKREKTGTDLSLIRSSLSANLVCSSLNDSEVEALANAVQFFTFTKGDIVTKQGE 165
+ + K KT ++ + ++ N++ S L+D+E + +A+ +F G+ V +QG+
Sbjct 113 YVRKVIPKDYKT---MAALAKAIEKNVLFSHLDDNERSDIFDAMFSVSFIAGETVIQQGD 169
Query 166 NGSYFFIVHSGEFEVIVNDKVVNKIVEGQAFGEISLIHNSARTASIKTLSEKAALWGVQR 225
G F+++ GE +V VN++ + EG +FGE++LI+ + R A++K + LWG+ R
Sbjct 170 EGDNFYVIDQGETDVYVNNEWATSVGEGGSFGELALIYGTPRAATVKAKT-NVKLWGIDR 228
Query 226 QVFRETLKQLSSRNFAENRQFLASVEFFQMLTEAQKNVITNALVVQSFRAGQPIVKEGEK 285
+R L + R +FL+ V + L + ++ + +AL F GQ IV +GE
Sbjct 229 DSYRRILMGSTLRKRKMYEEFLSKVSILESLDKWERLTVADALEPVQFEDGQKIVVQGEP 288
Query 286 GDILYILKSGKARV----SINGKDVRL--LQKGDYFGERALLYDEPRSATIIAEEETVCV 339
GD +I+ G A V S N + V + L DYFGE ALL + PR+AT++A CV
Sbjct 289 GDEFFIILEGSAAVLQRRSENEEFVEVGRLGPSDYFGEIALLMNRPRAATVVARGPLKCV 348
Query 340 SIGR 343
+ R
Sbjct 349 KLDR 352
Score = 61.2 bits (147), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 54/93 (58%), Gaps = 0/93 (0%)
Query 252 FFQMLTEAQKNVITNALVVQSFRAGQPIVKEGEKGDILYILKSGKARVSINGKDVRLLQK 311
F L + +++ I +A+ SF AG+ ++++G++GD Y++ G+ V +N + + +
Sbjct 137 LFSHLDDNERSDIFDAMFSVSFIAGETVIQQGDEGDNFYVIDQGETDVYVNNEWATSVGE 196
Query 312 GDYFGERALLYDEPRSATIIAEEETVCVSIGRD 344
G FGE AL+Y PR+AT+ A+ I RD
Sbjct 197 GGSFGELALIYGTPRAATVKAKTNVKLWGIDRD 229
> 7296360
Length=299
Score = 129 bits (324), Expect = 8e-30, Method: Compositional matrix adjust.
Identities = 74/230 (32%), Positives = 128/230 (55%), Gaps = 8/230 (3%)
Query 121 LSLIRSSLSANLVCSSLNDSEVEALANAVQFFTFTKGDIVTKQGENGSYFFIVHSGEFEV 180
++ + +++ N++ + L++SE + +A+ G+ + +QG+ G F+++ GE +V
Sbjct 40 MNALSKAIAKNVLFAHLDESERSDIFDAMFPVNHIAGENIIQQGDEGDNFYVIDVGEVDV 99
Query 181 IVNDKVVNKIVEGQAFGEISLIHNSARTASIKTLSEKAALWGVQRQVFRETLKQLSSRNF 240
VN ++V I EG +FGE++LI+ + R A+++ ++ LWG+ R +R L + R
Sbjct 100 FVNSELVTTISEGGSFGELALIYGTPRAATVRAKTD-VKLWGIDRDSYRRILMGSTIRKR 158
Query 241 AENRQFLASVEFFQMLTEAQKNVITNALVVQSFRAGQPIVKEGEKGDILYILKSGKARV- 299
+FL+ V + L + ++ + ++L SF G+ IVK+G GD YI+ G A V
Sbjct 159 KMYEEFLSRVSILESLDKWERLTVADSLETCSFDDGETIVKQGAAGDDFYIILEGCAVVL 218
Query 300 ---SINGKD---VRLLQKGDYFGERALLYDEPRSATIIAEEETVCVSIGR 343
S G+D V L DYFGE ALL D PR+AT++A CV + R
Sbjct 219 QQRSEQGEDPAEVGRLGSSDYFGEIALLLDRPRAATVVARGPLKCVKLDR 268
Score = 61.2 bits (147), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 31/93 (33%), Positives = 56/93 (60%), Gaps = 0/93 (0%)
Query 252 FFQMLTEAQKNVITNALVVQSFRAGQPIVKEGEKGDILYILKSGKARVSINGKDVRLLQK 311
F L E++++ I +A+ + AG+ I+++G++GD Y++ G+ V +N + V + +
Sbjct 52 LFAHLDESERSDIFDAMFPVNHIAGENIIQQGDEGDNFYVIDVGEVDVFVNSELVTTISE 111
Query 312 GDYFGERALLYDEPRSATIIAEEETVCVSIGRD 344
G FGE AL+Y PR+AT+ A+ + I RD
Sbjct 112 GGSFGELALIYGTPRAATVRAKTDVKLWGIDRD 144
> YIL033c
Length=416
Score = 129 bits (324), Expect = 8e-30, Method: Compositional matrix adjust.
Identities = 73/231 (31%), Positives = 124/231 (53%), Gaps = 2/231 (0%)
Query 114 REKTGTDLSLIRSSLSANLVCSSLNDSEVEALANAVQFFTFTKGDIVTKQGENGSYFFIV 173
+EK+ L + S+ N + + L+ + N ++ + KG + KQG+ G YF++V
Sbjct 165 KEKSEQQLQRLEKSIRNNFLFNKLDSDSKRLVINCLEEKSVPKGATIIKQGDQGDYFYVV 224
Query 174 HSGEFEVIVNDKVVNKIVEGQAFGEISLIHNSARTASIKTLSEKAALWGVQRQVFRETLK 233
G + VND VN G +FGE++L++NS R A++ S+ LW + R FR+ L
Sbjct 225 EKGTVDFYVNDNKVNSSGPGSSFGELALMYNSPRAATVVATSD-CLLWALDRLTFRKILL 283
Query 234 QLSSRNFAENRQFLASVEFFQMLTEAQKNVITNALVVQSFRAGQPIVKEGEKGDILYILK 293
S + L S+ + LT + + +AL + ++ G+ I++EG++G+ Y+++
Sbjct 284 GSSFKKRLMYDDLLKSMPVLKSLTTYDRAKLADALDTKIYQPGETIIREGDQGENFYLIE 343
Query 294 SGKARVSINGKDV-RLLQKGDYFGERALLYDEPRSATIIAEEETVCVSIGR 343
G VS G+ V L+ DYFGE ALL D PR AT+ A + T ++G+
Sbjct 344 YGAVDVSKKGQGVINKLKDHDYFGEVALLNDLPRQATVTATKRTKVATLGK 394
> CE19898
Length=737
Score = 122 bits (307), Expect = 9e-28, Method: Compositional matrix adjust.
Identities = 72/233 (30%), Positives = 126/233 (54%), Gaps = 7/233 (3%)
Query 111 LEKREKTGTDLSLIRSSLSANLVCSSLNDSEVEALANAVQFFTFTKGDIVTKQGENGSYF 170
L+ KT +IR ++ N L ++ L N + G V ++GE G
Sbjct 136 LQHYNKTVGAKQMIRDAVQKNDFLKQLAKEQIIELVNCMYEMRARAGQWVIQEGEPGDRL 195
Query 171 FIVHSGEFEVIVNDKVVNKIVEGQAFGEISLIHNSARTASIKTLSEKAALWGVQRQVFRE 230
F+V GE +V ++ K+ G GE+++++N RTAS++ L++ LW + R VF+
Sbjct 196 FVVAEGELQVSREGALLGKMRAGTVMGELAILYNCTRTASVQALTD-VQLWVLDRSVFQM 254
Query 231 TLKQLSSRNFAENRQFLASVEFFQMLTEAQKNVITNALVVQSFRAGQPIVKEGEKGDILY 290
++L ++ FL V FQ L+E + + + + + + G I+++GEKGD +
Sbjct 255 ITQRLGMERHSQLMNFLTKVSIFQNLSEDRISKMADVMDQDYYDGGHYIIRQGEKGDAFF 314
Query 291 ILKSGKARVS--ING----KDVRLLQKGDYFGERALLYDEPRSATIIAEEETV 337
++ SG+ +V+ I G +++R+L +GD+FGERALL +E R+A IIA+ V
Sbjct 315 VINSGQVKVTQQIEGETEPREIRVLNQGDFFGERALLGEEVRTANIIAQAPGV 367
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/111 (22%), Positives = 54/111 (48%), Gaps = 6/111 (5%)
Query 133 VCSSLNDSEVEALANAVQFFTFTKGDIVTKQGENGSYFFIVHSGEFEVI------VNDKV 186
+ +L++ + +A+ + + G + +QGE G FF+++SG+ +V +
Sbjct 276 IFQNLSEDRISKMADVMDQDYYDGGHYIIRQGEKGDAFFVINSGQVKVTQQIEGETEPRE 335
Query 187 VNKIVEGQAFGEISLIHNSARTASIKTLSEKAALWGVQRQVFRETLKQLSS 237
+ + +G FGE +L+ RTA+I + + + R+ F + + L S
Sbjct 336 IRVLNQGDFFGERALLGEEVRTANIIAQAPGVEVLTLDRESFGKLIGDLES 386
> CE19897
Length=780
Score = 122 bits (307), Expect = 9e-28, Method: Compositional matrix adjust.
Identities = 72/233 (30%), Positives = 126/233 (54%), Gaps = 7/233 (3%)
Query 111 LEKREKTGTDLSLIRSSLSANLVCSSLNDSEVEALANAVQFFTFTKGDIVTKQGENGSYF 170
L+ KT +IR ++ N L ++ L N + G V ++GE G
Sbjct 179 LQHYNKTVGAKQMIRDAVQKNDFLKQLAKEQIIELVNCMYEMRARAGQWVIQEGEPGDRL 238
Query 171 FIVHSGEFEVIVNDKVVNKIVEGQAFGEISLIHNSARTASIKTLSEKAALWGVQRQVFRE 230
F+V GE +V ++ K+ G GE+++++N RTAS++ L++ LW + R VF+
Sbjct 239 FVVAEGELQVSREGALLGKMRAGTVMGELAILYNCTRTASVQALTD-VQLWVLDRSVFQM 297
Query 231 TLKQLSSRNFAENRQFLASVEFFQMLTEAQKNVITNALVVQSFRAGQPIVKEGEKGDILY 290
++L ++ FL V FQ L+E + + + + + + G I+++GEKGD +
Sbjct 298 ITQRLGMERHSQLMNFLTKVSIFQNLSEDRISKMADVMDQDYYDGGHYIIRQGEKGDAFF 357
Query 291 ILKSGKARVS--ING----KDVRLLQKGDYFGERALLYDEPRSATIIAEEETV 337
++ SG+ +V+ I G +++R+L +GD+FGERALL +E R+A IIA+ V
Sbjct 358 VINSGQVKVTQQIEGETEPREIRVLNQGDFFGERALLGEEVRTANIIAQAPGV 410
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/111 (22%), Positives = 54/111 (48%), Gaps = 6/111 (5%)
Query 133 VCSSLNDSEVEALANAVQFFTFTKGDIVTKQGENGSYFFIVHSGEFEVI------VNDKV 186
+ +L++ + +A+ + + G + +QGE G FF+++SG+ +V +
Sbjct 319 IFQNLSEDRISKMADVMDQDYYDGGHYIIRQGEKGDAFFVINSGQVKVTQQIEGETEPRE 378
Query 187 VNKIVEGQAFGEISLIHNSARTASIKTLSEKAALWGVQRQVFRETLKQLSS 237
+ + +G FGE +L+ RTA+I + + + R+ F + + L S
Sbjct 379 IRVLNQGDFFGERALLGEEVRTANIIAQAPGVEVLTLDRESFGKLIGDLES 429
> CE28749
Length=334
Score = 120 bits (302), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 76/272 (27%), Positives = 138/272 (50%), Gaps = 16/272 (5%)
Query 81 PPKPGGERK--VQKAIMQQDDTQAEDARLLSHLEKREKTGTDLSLIRSSLSANLVCSSLN 138
PPK G R+ + +++DDT+ + + R + S++ NL+ + L
Sbjct 43 PPKRSGGRRTGISAEPIKEDDTEYKKVVIPKDDATRRS-------LESAMRKNLLFAHLE 95
Query 139 DSEVEALANAVQFFTFTKGDIVTKQGENGSYFFIVHSGEFEVIVNDKVVNKIVEGQAFGE 198
+ E + + +A+ + G+ + +QGE G F+++ G +V VN + V I EG +FGE
Sbjct 96 EDEQKTMYDAMFPVEKSAGETIIEQGEEGDNFYVIDKGTVDVYVNHEYVLTINEGGSFGE 155
Query 199 ISLIHNSARTASIKTLSEKAALWGVQRQVFRETLKQLSSRNFAENRQFLASVEFFQMLTE 258
++LI+ + R A++ ++ LW + R +R L ++ +FL+ V+ L +
Sbjct 156 LALIYGTPRAATVIAKTD-VKLWAIDRLTYRRILMGSVTKKRKMYDEFLSKVQILADLDQ 214
Query 259 AQKNVITNALVVQSFRAGQPIVKEGEKGDILYILKSGKARVSINGKD------VRLLQKG 312
++ + +AL F G +V++G+ GD +I+ G+A V D V L
Sbjct 215 WERANVADALERCDFEPGTHVVEQGQPGDEFFIILEGEANVLQKRSDDAPFDVVGHLGMS 274
Query 313 DYFGERALLYDEPRSATIIAEEETVCVSIGRD 344
DYFGE ALL D PR+AT++A+ C+ + R+
Sbjct 275 DYFGEIALLLDRPRAATVVAKTHLKCIKLDRN 306
> CE04821
Length=376
Score = 120 bits (300), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 78/272 (28%), Positives = 140/272 (51%), Gaps = 16/272 (5%)
Query 81 PPKPGGERK--VQKAIMQQDDTQAEDARLLSHLEKREKTGTDLSLIRSSLSANLVCSSLN 138
PPK G R+ + +++DDT+ + + K + T L S++ NL+ + L
Sbjct 85 PPKRSGGRRTGISAEPIKEDDTEYKKVVI----PKDDATRRSLE---SAMRKNLLFAHLE 137
Query 139 DSEVEALANAVQFFTFTKGDIVTKQGENGSYFFIVHSGEFEVIVNDKVVNKIVEGQAFGE 198
+ E + + +A+ + G+ + +QGE G F+++ G +V VN + V I EG +FGE
Sbjct 138 EDEQKTMYDAMFPVEKSAGETIIEQGEEGDNFYVIDKGTVDVYVNHEYVLTINEGGSFGE 197
Query 199 ISLIHNSARTASIKTLSEKAALWGVQRQVFRETLKQLSSRNFAENRQFLASVEFFQMLTE 258
++LI+ + R A++ ++ LW + R +R L ++ +FL+ V+ L +
Sbjct 198 LALIYGTPRAATVIAKTD-VKLWAIDRLTYRRILMGSVTKKRKMYDEFLSKVQILADLDQ 256
Query 259 AQKNVITNALVVQSFRAGQPIVKEGEKGDILYILKSGKARVSINGKD------VRLLQKG 312
++ + +AL F G +V++G+ GD +I+ G+A V D V L
Sbjct 257 WERANVADALERCDFEPGTHVVEQGQPGDEFFIILEGEANVLQKRSDDAPFDVVGHLGMS 316
Query 313 DYFGERALLYDEPRSATIIAEEETVCVSIGRD 344
DYFGE ALL D PR+AT++A+ C+ + R+
Sbjct 317 DYFGEIALLLDRPRAATVVAKTHLKCIKLDRN 348
> Hs10835242
Length=686
Score = 118 bits (296), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 91/332 (27%), Positives = 163/332 (49%), Gaps = 16/332 (4%)
Query 26 EQQQQQQQQQEQQQQRQP---QHQQSQQQQQEPQQQQEQQQPPDRKTSQP--QQSDDAAA 80
+ Q Q++ E+ +QR + + Q+ E Q+ + + R +P QQ+ +A
Sbjct 6 DLQYALQEKIEELRQRDALIDELELELDQKDELIQKLQNELDKYRSVIRPATQQAQKQSA 65
Query 81 PPKPGGERKVQKAIMQQDDTQAEDARLLSH--LEKREKTGTDLSLIRSSLSANLVCSSLN 138
G R ++AI + A D + LSH L K+ LI+ ++ N +L
Sbjct 66 STLQGEPRTKRQAISAE--PTAFDIQDLSHVTLPFYPKSPQSKDLIKEAILDNDFMKNLE 123
Query 139 DSEVEALANAVQFFTFTKGDIVTKQGENGSYFFIVHSGEFEVIVNDKVVNKIVEGQAFGE 198
S+++ + + + + K + K+G+ GS +++ G+ EV + + G+ FGE
Sbjct 124 LSQIQEIVDCMYPVEYGKDSCIIKEGDVGSLVYVMEDGKVEVTKEGVKLCTMGPGKVFGE 183
Query 199 ISLIHNSARTASIKTLSEKAALWGVQRQVFRETLKQLSSRNFAENRQFLASVEFFQMLTE 258
+++++N RTA++KTL LW + RQ F+ + + E +FL SV FQ L E
Sbjct 184 LAILYNCTRTATVKTLV-NVKLWAIDRQCFQTIMMRTGLIKHTEYMEFLKSVPTFQSLPE 242
Query 259 AQKNVITNALVVQSFRAGQPIVKEGEKGDILYILKSGKARV------SINGKDVRLLQKG 312
+ + + L + G+ I+++G +GD +I+ G V S + +R L KG
Sbjct 243 EILSKLADVLEETHYENGEYIIRQGARGDTFFIISKGTVNVTREDSPSEDPVFLRTLGKG 302
Query 313 DYFGERALLYDEPRSATIIAEEETVCVSIGRD 344
D+FGE+AL ++ R+A +IA E C+ I RD
Sbjct 303 DWFGEKALQGEDVRTANVIAAEAVTCLVIDRD 334
> 7303815
Length=411
Score = 115 bits (289), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 58/180 (32%), Positives = 101/180 (56%), Gaps = 5/180 (2%)
Query 157 GDIVTKQGENGSYFFIVHSGEFEVIVNDKVVNKIVEGQAFGEISLIHNSARTASIKTLSE 216
GD + +QG++G F+++ SG ++V +NDK +N FGE++L++N R A+++
Sbjct 148 GDFIIRQGDDGDNFYVIESGVYKVYINDKHINTYNHTGLFGELALLYNMPRAATVQA-ET 206
Query 217 KAALWGVQRQVFRETLKQLSSRNFAENRQFLASVEFFQMLTEAQKNVITNALVVQSFRAG 276
LW + RQ FR L + + R + L SV + L ++ + +ALV +S+ G
Sbjct 207 SGLLWAMDRQTFRRILLKSAFRKRKMYEELLNSVPMLKALQNYERMNLADALVSKSYDNG 266
Query 277 QPIVKEGEKGDILYILKSGKARVSINGKDVRL----LQKGDYFGERALLYDEPRSATIIA 332
+ I+K+G+ D +Y ++ G V ++ D + L KG YFGE AL+ PR+A++ A
Sbjct 267 ERIIKQGDAADGMYFIEEGTVSVRMDQDDAEVEISQLGKGQYFGELALVTHRPRAASVYA 326
Score = 64.3 bits (155), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 32/95 (33%), Positives = 55/95 (57%), Gaps = 0/95 (0%)
Query 249 SVEFFQMLTEAQKNVITNALVVQSFRAGQPIVKEGEKGDILYILKSGKARVSINGKDVRL 308
+V F+ L + Q N + +A+ + + G I+++G+ GD Y+++SG +V IN K +
Sbjct 121 NVLLFRSLEKEQMNQVLDAMFERKVQPGDFIIRQGDDGDNFYVIESGVYKVYINDKHINT 180
Query 309 LQKGDYFGERALLYDEPRSATIIAEEETVCVSIGR 343
FGE ALLY+ PR+AT+ AE + ++ R
Sbjct 181 YNHTGLFGELALLYNMPRAATVQAETSGLLWAMDR 215
> 7297611
Length=1003
Score = 106 bits (264), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 67/220 (30%), Positives = 120/220 (54%), Gaps = 11/220 (5%)
Query 122 SLIRSSLSANLVCSSLNDSE-VEALANAVQFFTFTKGDIVTKQGENGSYFFIVHSGEFEV 180
+LIR+++ N ++L D E E + NA+ ++ K ++ ++ E GS ++ G+++V
Sbjct 420 NLIRTAIERNDFLNNLMDKERKEMVINAMAPASYRKHSLIIREHEEGSEIYVSAEGQYDV 479
Query 181 IVNDKVVNKIVEGQAFGEISLIHNSARTASIKTLSEKAALWGVQRQVFRETLKQLSSRNF 240
I ++V FGE+++++N+ R ASI+ ++ A +W + R+ FR ++ SR
Sbjct 480 IRGGQLVASFGPATVFGELAILYNAPRQASIEAATD-ARVWKIARETFRAIMQISGSRER 538
Query 241 AENRQFLASVEFFQMLTEAQKNVITNALVVQSFRAGQPIVKEGEKGDILYILKSGKARVS 300
EN QFL S F Q ++ + + L + + IV+EGE G+ YI++ G V+
Sbjct 539 EENLQFLRSAPFLQEFDQSLLLKVVDLLQRKFYETDSCIVREGELGNEFYIIRCGT--VT 596
Query 301 INGKD-------VRLLQKGDYFGERALLYDEPRSATIIAE 333
I D V ++GDYFGE+ALL + R A++ A+
Sbjct 597 IKKLDEQQQEQIVANRKRGDYFGEQALLNADVRQASVYAD 636
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/90 (30%), Positives = 47/90 (52%), Gaps = 0/90 (0%)
Query 255 MLTEAQKNVITNALVVQSFRAGQPIVKEGEKGDILYILKSGKARVSINGKDVRLLQKGDY 314
++ + +K ++ NA+ S+R I++E E+G +Y+ G+ V G+ V
Sbjct 435 LMDKERKEMVINAMAPASYRKHSLIIREHEEGSEIYVSAEGQYDVIRGGQLVASFGPATV 494
Query 315 FGERALLYDEPRSATIIAEEETVCVSIGRD 344
FGE A+LY+ PR A+I A + I R+
Sbjct 495 FGELAILYNAPRQASIEAATDARVWKIARE 524
Score = 35.0 bits (79), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 48/106 (45%), Gaps = 13/106 (12%)
Query 160 VTKQGENGSYFFIVHSGEFEVIVNDK-----VVNKIVEGQAFGEISLIHNSARTASIKTL 214
+ ++GE G+ F+I+ G + D+ +V G FGE +L++ R AS+
Sbjct 577 IVREGELGNEFYIIRCGTVTIKKLDEQQQEQIVANRKRGDYFGEQALLNADVRQASVYAD 636
Query 215 SEKAALWGVQRQVFRE---TLKQL-----SSRNFAENRQFLASVEF 252
+ + + R+ F T+KQL S RN R S+EF
Sbjct 637 APGTEVLMLDREAFISYLGTIKQLREKPSSQRNDTSGRSSTKSLEF 682
> Hs4758958
Length=404
Score = 98.2 bits (243), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 53/198 (26%), Positives = 106/198 (53%), Gaps = 17/198 (8%)
Query 160 VTKQGENGSYFFIVHSGEFEVIVND----KVVNKIVEGQAFGEISLIHNSARTASIKTLS 215
V QG++G F+++ G ++++V + V + +FGE++L++N+ R A+I S
Sbjct 166 VIDQGDDGDNFYVIERGTYDILVTKDNQTRSVGQYDNRGSFGELALMYNTPRAATIVATS 225
Query 216 EKAALWGVQRQVFRETLKQLSSRNFAENRQFLASVEFFQMLTEAQKNVITNALVVQSFRA 275
E +LWG+ R FR + + +++ F+ SV + L +++ I + + + ++
Sbjct 226 E-GSLWGLDRVTFRRIIVKNNAKKRKMFESFIESVPLLKSLEVSERMKIVDVIGEKIYKD 284
Query 276 GQPIVKEGEKGDILYILKSGKARVSINGK------------DVRLLQKGDYFGERALLYD 323
G+ I+ +GEK D YI++SG+ + I + ++ KG YFGE AL+ +
Sbjct 285 GERIITQGEKADSFYIIESGEVSILIRSRTKSNKDGGNQEVEIARCHKGQYFGELALVTN 344
Query 324 EPRSATIIAEEETVCVSI 341
+PR+A+ A + C+ +
Sbjct 345 KPRAASAYAVGDVKCLVM 362
> Hs4506065
Length=418
Score = 94.7 bits (234), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 51/200 (25%), Positives = 108/200 (54%), Gaps = 16/200 (8%)
Query 157 GDIVTKQGENGSYFFIVHSGEFEVIVNDKVVNKIVEGQ----AFGEISLIHNSARTASIK 212
G+ V QG++G F+++ G F++ V V + V +FGE++L++N+ R A+I
Sbjct 178 GEHVIDQGDDGDNFYVIDRGTFDIYVKCDGVGRCVGNYDNRGSFGELALMYNTPRAATI- 236
Query 213 TLSEKAALWGVQRQVFRETLKQLSSRNFAENRQFLASVEFFQMLTEAQKNVITNALVVQS 272
T + ALWG+ R FR + + +++ F+ S+ F + L +++ + + + +
Sbjct 237 TATSPGALWGLDRVTFRRIIVKNNAKKRKMYESFIESLPFLKSLEFSERLKVVDVIGTKV 296
Query 273 FRAGQPIVKEGEKGDILYILKSGKARVSINGK-----------DVRLLQKGDYFGERALL 321
+ G+ I+ +G+ D +I++SG+ ++++ K ++ +G YFGE AL+
Sbjct 297 YNDGEQIIAQGDSADSFFIVESGEVKITMKRKGKSEVEENGAVEMPRCSRGQYFGELALV 356
Query 322 YDEPRSATIIAEEETVCVSI 341
++PR+A+ A C+++
Sbjct 357 TNKPRAASAHAIGTVKCLAM 376
> SPAC8C9.03
Length=412
Score = 94.0 bits (232), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 67/228 (29%), Positives = 120/228 (52%), Gaps = 17/228 (7%)
Query 120 DLSLIRSSLSANLVCSSLNDSEVEALANAVQFFTFTKGDI-VTKQGENGSYFFIVHSGEF 178
DL ++ S++ N + +L++ + NA+ + + V QG G YF+IV GEF
Sbjct 130 DLKRLKRSVAGNFLFKNLDEEHYNEVLNAMTEKRIGEAGVAVIVQGAVGDYFYIVEQGEF 189
Query 179 EV-------IVNDKV--------VNKIVEGQAFGEISLIHNSARTASIKTLSEKAALWGV 223
+V I ++V + I G+ FGE++L++N+ R AS+ + + ++ +
Sbjct 190 DVYKRPELNITPEEVLSSGYGNYITTISPGEYFGELALMYNAPRAASVVSKTPNNVIYAL 249
Query 224 QRQVFRETLKQLSSRNFAENRQFLASVEFFQMLTEAQKNVITNALVVQSFRAGQPIVKEG 283
R FR + + + R L V L + Q+ I +AL ++AG ++++G
Sbjct 250 DRTSFRRIVFENAYRQRMLYESLLEEVPILSSLDKYQRQKIADALQTVVYQAGSIVIRQG 309
Query 284 EKGDILYILKSGKARVSINGKDVRL-LQKGDYFGERALLYDEPRSATI 330
+ G+ Y+++ G+A V NGK V + L KGDYFGE AL+++ R+AT+
Sbjct 310 DIGNQFYLIEDGEAEVVKNGKGVVVTLTKGDYFGELALIHETVRNATV 357
> CE03979
Length=581
Score = 67.4 bits (163), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 29/111 (26%), Positives = 67/111 (60%), Gaps = 1/111 (0%)
Query 124 IRSSLSANLVCSSLNDSEVEALANAVQFFTFTKGDIVTKQGENGSYFFIVHSGEFEVIVN 183
I ++L N +L+ +++E +++A+ G I+ +QG+ GS +++ G+ +V+ +
Sbjct 27 IGNALRLNSFLRNLDATQIEKISSAMYPVEVPAGAIIIRQGDLGSIMYVIQEGKVQVVKD 86
Query 184 DKVVNKIVEGQAFGEISLIHNSARTASIKTLSEKAALWGVQRQVFRETLKQ 234
++ V + +G FGE++++H+ RTA+++ + E LW ++R VF + +
Sbjct 87 NRFVRTMEDGALFGELAILHHCERTATVRAI-ESCHLWAIERNVFHAIMME 136
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 53/93 (56%), Gaps = 0/93 (0%)
Query 252 FFQMLTEAQKNVITNALVVQSFRAGQPIVKEGEKGDILYILKSGKARVSINGKDVRLLQK 311
F + L Q I++A+ AG I+++G+ G I+Y+++ GK +V + + VR ++
Sbjct 36 FLRNLDATQIEKISSAMYPVEVPAGAIIIRQGDLGSIMYVIQEGKVQVVKDNRFVRTMED 95
Query 312 GDYFGERALLYDEPRSATIIAEEETVCVSIGRD 344
G FGE A+L+ R+AT+ A E +I R+
Sbjct 96 GALFGELAILHHCERTATVRAIESCHLWAIERN 128
> ECU08g0930
Length=312
Score = 66.2 bits (160), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 39/121 (32%), Positives = 67/121 (55%), Gaps = 4/121 (3%)
Query 108 LSHLEKREKTGTDLSLIRSSLSANLVCSSLNDSEVEALANAVQFFTFTKGDIVTKQGENG 167
L++ K E+T + + S L +++ LN + L + + +G V +GE G
Sbjct 96 LNYYPKDEET---IRFLSSILVSDIPFGFLNPEQKTRLIESTELIEIKRGTFVMHEGEIG 152
Query 168 SYFFIVHSGEFEVIVNDKVVNKIVEGQAFGEISLIHNSARTASIKTLSEKAALWGVQRQV 227
S +IV SGEFEV ++ K+ +G FGEI+L+HN RTA++K +++ +W V++
Sbjct 153 SQMYIVASGEFEVTKGGTLLRKLTKGCFFGEIALLHNIPRTATVKAVTD-GKVWVVEQTS 211
Query 228 F 228
F
Sbjct 212 F 212
Score = 54.7 bits (130), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 34/95 (35%), Positives = 53/95 (55%), Gaps = 4/95 (4%)
Query 242 ENRQFLASVEF----FQMLTEAQKNVITNALVVQSFRAGQPIVKEGEKGDILYILKSGKA 297
E +FL+S+ F L QK + + + + G ++ EGE G +YI+ SG+
Sbjct 104 ETIRFLSSILVSDIPFGFLNPEQKTRLIESTELIEIKRGTFVMHEGEIGSQMYIVASGEF 163
Query 298 RVSINGKDVRLLQKGDYFGERALLYDEPRSATIIA 332
V+ G +R L KG +FGE ALL++ PR+AT+ A
Sbjct 164 EVTKGGTLLRKLTKGCFFGEIALLHNIPRTATVKA 198
> CE23992
Length=1234
Score = 58.2 bits (139), Expect = 2e-08, Method: Composition-based stats.
Identities = 32/81 (39%), Positives = 51/81 (62%), Gaps = 1/81 (1%)
Query 256 LTEAQKNVITNALVVQSF-RAGQPIVKEGEKGDILYILKSGKARVSINGKDVRLLQKGDY 314
L+ K ++N + V+ + AG + ++GE G YI+ G V++NGK V LL++GD
Sbjct 575 LSTMVKRQLSNFVKVEQYVHAGSVVFRQGEIGVYWYIVLKGAVEVNVNGKIVCLLREGDD 634
Query 315 FGERALLYDEPRSATIIAEEE 335
FG+ AL+ D PR+ATI+ E+
Sbjct 635 FGKLALVNDLPRAATIVTYED 655
Score = 57.4 bits (137), Expect = 5e-08, Method: Composition-based stats.
Identities = 32/100 (32%), Positives = 56/100 (56%), Gaps = 5/100 (5%)
Query 157 GDIVTKQGENGSYFFIVHSGEFEVIVNDKVVNKIVEGQAFGEISLIHNSARTASIKTLSE 216
G +V +QGE G Y++IV G EV VN K+V + EG FG+++L+++ R A+I T +
Sbjct 596 GSVVFRQGEIGVYWYIVLKGAVEVNVNGKIVCLLREGDDFGKLALVNDLPRAATIVTYED 655
Query 217 KAALWGVQRQVFRETLKQLSS-----RNFAENRQFLASVE 251
+ V + F + L Q+ + +++ E+ L V+
Sbjct 656 DSMFLVVDKHHFNQILHQVEANTVRLKDYGEDVLVLEKVD 695
> Hs20070215
Length=881
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 42/169 (24%), Positives = 83/169 (49%), Gaps = 13/169 (7%)
Query 82 PKPGGERKVQK------AIMQQDDTQAEDARLLSHLEKR--EKTGTDLSLIRSSLSANLV 133
P+P G ++++ A++ Q + DA L L K ++T +L LI L
Sbjct 147 PEPVGTHEMEEELAEAVALLSQ---RGPDALLTVALRKPPGQRTDEELDLIFEELLHIKA 203
Query 134 CSSLNDSEVEALANAVQFFTFTK-GDIVTKQGENGSYFFIVHSGEFEVIVNDK-VVNKIV 191
+ L++S LA + F +K G ++ QG+ G+ ++I+ G V+ + K +V +
Sbjct 204 VAHLSNSVKRELAAVLLFEPHSKAGTVLFSQGDKGTSWYIIWKGSVNVVTHGKGLVTTLH 263
Query 192 EGQAFGEISLIHNSARTASIKTLSEKAALWGVQRQVFRETLKQLSSRNF 240
EG FG+++L++++ R A+I + V +Q F +K + ++
Sbjct 264 EGDDFGQLALVNDAPRAATIILREDNCHFLRVDKQDFNRIIKDVEAKTM 312
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/65 (41%), Positives = 38/65 (58%), Gaps = 1/65 (1%)
Query 274 RAGQPIVKEGEKGDILYILKSGKARVSINGKD-VRLLQKGDYFGERALLYDEPRSATIIA 332
+AG + +G+KG YI+ G V +GK V L +GD FG+ AL+ D PR+ATII
Sbjct 226 KAGTVLFSQGDKGTSWYIIWKGSVNVVTHGKGLVTTLHEGDDFGQLALVNDAPRAATIIL 285
Query 333 EEETV 337
E+
Sbjct 286 REDNC 290
> 7291428
Length=1991
Score = 30.4 bits (67), Expect = 5.2, Method: Composition-based stats.
Identities = 9/31 (29%), Positives = 26/31 (83%), Gaps = 0/31 (0%)
Query 26 EQQQQQQQQQEQQQQRQPQHQQSQQQQQEPQ 56
EQ++++Q+++EQ+++ Q + +Q +++Q+E +
Sbjct 1422 EQREKEQREKEQREKEQREREQREKEQREKE 1452
Lambda K H
0.311 0.126 0.337
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8270168410
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40