bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0336_orf2
Length=153
Score E
Sequences producing significant alignments: (Bits) Value
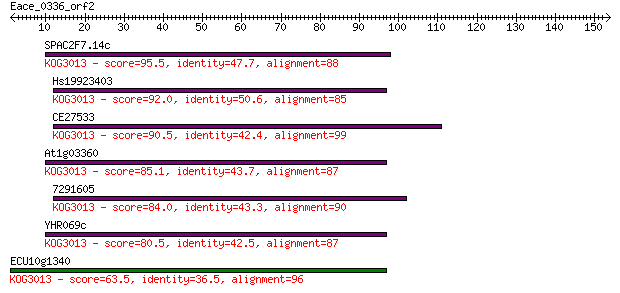
SPAC2F7.14c 95.5 3e-20
Hs19923403 92.0 4e-19
CE27533 90.5 1e-18
At1g03360 85.1 5e-17
7291605 84.0 1e-16
YHR069c 80.5 1e-15
ECU10g1340 63.5 1e-10
> SPAC2F7.14c
Length=329
Score = 95.5 bits (236), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 42/88 (47%), Positives = 61/88 (69%), Gaps = 0/88 (0%)
Query 10 VQRRRDDADVLIMRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQNGIGLRVSPQL 69
+QRR+ + D L MRS FQ DLL EVQ+ +DG++ +HTRS +YG+L+NG+ L+V P L
Sbjct 150 IQRRKLETDELQMRSFFQEGDLLVAEVQQYFSDGSVSIHTRSLKYGKLRNGVFLKVPPAL 209
Query 70 IKRQSKHIGHLQCGLQLILGVNGFLWIS 97
+ R H L G+ +IL VNG++W+S
Sbjct 210 VVRSKSHAYALAGGVDIILSVNGYVWVS 237
> Hs19923403
Length=293
Score = 92.0 bits (227), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 43/85 (50%), Positives = 54/85 (63%), Gaps = 0/85 (0%)
Query 12 RRRDDADVLIMRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQNGIGLRVSPQLIK 71
RRR D L MR Q DL+ EVQ +DG + LHTRS +YG+L G+ ++VSP L+K
Sbjct 121 RRRSAEDELAMRGFLQEGDLISAEVQAVFSDGAVSLHTRSLKYGKLGQGVLVQVSPSLVK 180
Query 72 RQSKHIGHLQCGLQLILGVNGFLWI 96
RQ H L CG +ILG NGF+WI
Sbjct 181 RQKTHFHDLPCGASVILGNNGFIWI 205
> CE27533
Length=303
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 42/101 (41%), Positives = 63/101 (62%), Gaps = 2/101 (1%)
Query 12 RRRDDADVLIMRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQNGIGLRVSPQLIK 71
RR+D D M ++ +L+C EVQ+ Q+DGT++LHTR+ +YG+LQ GI ++V P LIK
Sbjct 127 RRKDVEDEEKMSEFLKNGELICAEVQQVQHDGTLMLHTRNNKYGKLQQGILIKVPPHLIK 186
Query 72 RQSKHIGHLQCGLQLILGVNGFLWI--SVPGGAAGGSGDGV 110
+ KH L G+ +I+G NG +W+ S+P G V
Sbjct 187 KSKKHFHTLPYGMAVIIGCNGSVWVTPSLPETTLEEDGSHV 227
> At1g03360
Length=322
Score = 85.1 bits (209), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 38/88 (43%), Positives = 59/88 (67%), Gaps = 1/88 (1%)
Query 10 VQRRRDDADVLIMRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQNGIGLRVSPQL 69
+QRRR D L MR++F D++C EV+ Q+DG++ L RS +YG+L+ G L+V P L
Sbjct 132 IQRRRTSVDELNMRNIFVEHDVVCAEVRNFQHDGSLQLQARSQKYGKLEKGQLLKVDPYL 191
Query 70 IKRQSKHIGHLQC-GLQLILGVNGFLWI 96
+KR H +++ G+ LI+G NGF+W+
Sbjct 192 VKRSKHHFHYVESLGIDLIIGCNGFIWV 219
> 7291605
Length=298
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 39/90 (43%), Positives = 55/90 (61%), Gaps = 0/90 (0%)
Query 12 RRRDDADVLIMRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQNGIGLRVSPQLIK 71
RRR D +MR DL+ EVQ +G++ L+TRS +YG+L GI ++V P L+K
Sbjct 124 RRRSAEDEQMMRRYLDEGDLISAEVQNIFEEGSLSLYTRSLKYGKLSQGILVKVFPALVK 183
Query 72 RQSKHIGHLQCGLQLILGVNGFLWISVPGG 101
R+ H +L CG +ILG NG++WIS G
Sbjct 184 RRKMHFHNLPCGASVILGNNGYIWISPTKG 213
> YHR069c
Length=359
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 37/87 (42%), Positives = 55/87 (63%), Gaps = 0/87 (0%)
Query 10 VQRRRDDADVLIMRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQNGIGLRVSPQL 69
+ RR+ ++D L MRS + DLL EVQ DG+ LHTRS +YG+L+NG+ +V L
Sbjct 147 ILRRKSESDELQMRSFLKEGDLLNAEVQSLFQDGSASLHTRSLKYGKLRNGMFCQVPSSL 206
Query 70 IKRQSKHIGHLQCGLQLILGVNGFLWI 96
I R H +L + ++LGVNG++W+
Sbjct 207 IVRAKNHTHNLPGNITVVLGVNGYIWL 233
> ECU10g1340
Length=223
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 35/98 (35%), Positives = 56/98 (57%), Gaps = 3/98 (3%)
Query 1 LSVAAVCL--DVQRRRDDADVLIMRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQ 58
L ++A+ L VQRR+ ++D + MR F+ DL+ EVQK +G LHTRS +YG+L
Sbjct 87 LGLSAINLPGTVQRRKQESDEISMRDFFEIDDLVVSEVQKVGRNGVAALHTRSDKYGKLG 146
Query 59 NGIGLRVSPQLIKRQSKHIGHLQCGLQLILGVNGFLWI 96
G+ L P + K +++I+G NG++W+
Sbjct 147 PGL-LVFVPHFLLEPLKTRFLSNGSVEVIVGCNGYIWV 183
Lambda K H
0.322 0.135 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1961355924
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40