bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0317_orf2
Length=223
Score E
Sequences producing significant alignments: (Bits) Value
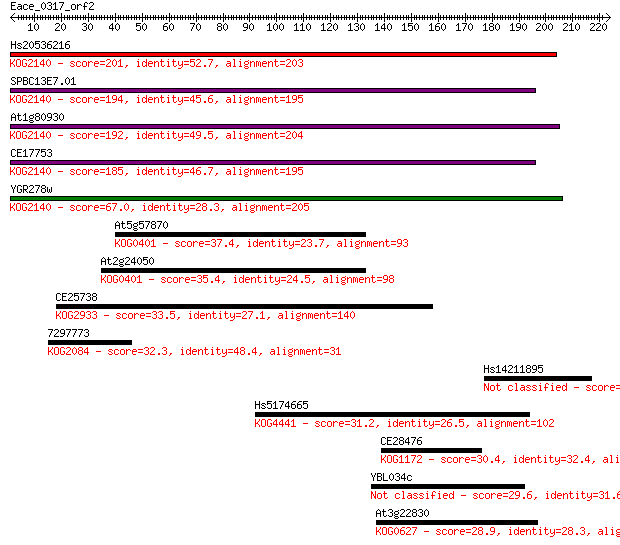
Hs20536216 201 1e-51
SPBC13E7.01 194 1e-49
At1g80930 192 4e-49
CE17753 185 5e-47
YGR278w 67.0 3e-11
At5g57870 37.4 0.026
At2g24050 35.4 0.091
CE25738 33.5 0.39
7297773 32.3 0.84
Hs14211895 31.6 1.5
Hs5174665 31.2 1.8
CE28476 30.4 3.4
YBL034c 29.6 4.8
At3g22830 28.9 9.5
> Hs20536216
Length=908
Score = 201 bits (511), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 107/203 (52%), Positives = 147/203 (72%), Gaps = 3/203 (1%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
AAL++I+NSK P IGEL++KRLIL FR+ YRR+DK +CL +F AHL+NQ +AHE+L L
Sbjct 216 AALVAIINSKFPQIGELILKRLILNFRKGYRRNDKQLCLTASKFVAHLINQNVAHEVLCL 275
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
++ LLL++PT+DSVEV IGFLK G L V G +A FERLR IL E E DK+ QY
Sbjct 276 EMLTLLLERPTDDSVEVAIGFLKECGLKLTQVSPRGINAIFERLRNILHESEIDKRVQYM 335
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
IE ++ +R++GFKD P ++E LDLVE +D+ TH + + D E++LN+FK DP+ +
Sbjct 336 IEVMFAVRKDGFKDHPIILEGLDLVEEDDQFTHMLPLED-DYNPEDVLNVFK-MDPN-FM 392
Query 181 EDEKKWSALSKEILGEDSTDSDA 203
E+E+K+ A+ KEIL E TDS+
Sbjct 393 ENEEKYKAIKKEILDEGDTDSNT 415
> SPBC13E7.01
Length=628
Score = 194 bits (493), Expect = 1e-49, Method: Compositional matrix adjust.
Identities = 89/195 (45%), Positives = 141/195 (72%), Gaps = 3/195 (1%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
AA+ +++N+K P IGEL++ RLI+QFR++++R+DK +C+ F AHL+NQ+IAHE++ L
Sbjct 177 AAMTAVINTKFPQIGELLLTRLIVQFRKSFQRNDKSMCISSSSFIAHLINQKIAHEIVGL 236
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
Q+ +LL++PTNDS+E+ + L+ +G L +V ++ FER R IL EG+ +++TQ+
Sbjct 237 QILAVLLERPTNDSIEIAVMLLREIGAYLAEVSTRAYNGVFERFRTILHEGQLERRTQFI 296
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
IE L+ R++ FK+ P + ++LDLVE ED+ITH + +D ++ +E L IF DPD YE
Sbjct 297 IEVLFQTRKDKFKNNPTIPQELDLVEEEDQITHYIS-LDDNLDVQESLGIFH-YDPD-YE 353
Query 181 EDEKKWSALSKEILG 195
E+EKK+ A+ EILG
Sbjct 354 ENEKKYDAIKHEILG 368
> At1g80930
Length=900
Score = 192 bits (489), Expect = 4e-49, Method: Compositional matrix adjust.
Identities = 101/205 (49%), Positives = 145/205 (70%), Gaps = 6/205 (2%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
AAL++++N+K P++ EL++KR++LQ +RAY+R+DK L V+F AHLVNQ++A E++AL
Sbjct 416 AALVAVINAKFPEVAELLLKRVVLQLKRAYKRNDKPQLLAAVKFIAHLVNQQVAEEIIAL 475
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
+L +LL PT+DSVEV +GF+ G +L+DV G + FER R IL EGE DK+ QY
Sbjct 476 ELVTILLGDPTDDSVEVAVGFVTECGAMLQDVSPRGLNGIFERFRGILHEGEIDKRVQYL 535
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
IE+L+ R+ F+ P V +LDL VE+K +H++ +D I E L+IFKP DPD +
Sbjct 536 IESLFATRKAKFQGHPAVRPELDL--VEEKYSHDLS-LDHEIDPETALDIFKP-DPD-FV 590
Query 181 EDEKKWSALSKEILG-EDSTDSDAS 204
E+EKK+ AL KE+LG E+S D D S
Sbjct 591 ENEKKYEALKKELLGDEESEDEDGS 615
> CE17753
Length=897
Score = 185 bits (470), Expect = 5e-47, Method: Compositional matrix adjust.
Identities = 91/200 (45%), Positives = 143/200 (71%), Gaps = 8/200 (4%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
AAL +++NSK P +GEL+++RLI+QF+R++RR+D+ V + ++F AHL+NQ++AHE+LAL
Sbjct 247 AALAAVINSKFPHVGELLLRRLIVQFKRSFRRNDRGVTVNVIKFIAHLINQQVAHEVLAL 306
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGET-----DK 115
++ L+L++PT+DSVEV I FLK G L ++ ++ ++RLRAIL E E D+
Sbjct 307 EIMILMLEEPTDDSVEVAIAFLKECGAKLLEIAPAALNSVYDRLRAILMETERSENALDR 366
Query 116 KTQYSIEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQD 175
+ QY IE IR++ F +P VIEDLDL+E ED+I H +++ D ++ E LN+FK D
Sbjct 367 RIQYMIETAMQIRKDKFAAYPAVIEDLDLIEEEDQIIHTLNLED-AVDPENGLNVFK-LD 424
Query 176 PDEYEEDEKKWSALSKEILG 195
P E+E++E+ + + KEI+G
Sbjct 425 P-EFEKNEEVYEEIRKEIIG 443
> YGR278w
Length=577
Score = 67.0 bits (162), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 58/205 (28%), Positives = 106/205 (51%), Gaps = 7/205 (3%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
+AL++++NS +PDIGE + K L+L F + + R D V C ++ + L + HE++ L
Sbjct 77 SALIALLNSDIPDIGETLAKELMLMFVQQFNRKDYVSCGNILQCLSILFLYDVIHEIVIL 136
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
Q+ LLL++ +S+ + I +K G L V + D +E+LR ILQ E + S
Sbjct 137 QILLLLLEK---NSLRLVIAVMKICGWKLALVSKKTHDMIWEKLRYILQTQELSSTLRES 193
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
+E L++IR+ +K + LD TH + D +E+ N K ++ +
Sbjct 194 LETLFEIRQKDYKSGSQGLFILDPTSYTVH-THSYIVSDEDEANKELGNFEKCEN---FN 249
Query 181 EDEKKWSALSKEILGEDSTDSDASS 205
E + L +++L +++D++ S
Sbjct 250 ELTMAFDTLRQKLLINNTSDTNEGS 274
> At5g57870
Length=780
Score = 37.4 bits (85), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 22/100 (22%), Positives = 49/100 (49%), Gaps = 7/100 (7%)
Query 40 GCVEFFAHLVNQRIAHELLALQLCELLLQQ-----PTNDSVEVCIGFLKAVGQVLEDVCR 94
G + L+ Q++ E + + + LL P ++VE F K +G+ L+ +
Sbjct 343 GNIRLIGELLKQKMVPEKIVHHIVQELLGADEKVCPAEENVEAICHFFKTIGKQLDGNVK 402
Query 95 GGF--DAAFERLRAILQEGETDKKTQYSIEALWDIRRNGF 132
D F+RL+A+ + + + + ++ ++ + D+R NG+
Sbjct 403 SKRINDVYFKRLQALSKNPQLELRLRFMVQNIIDMRSNGW 442
> At2g24050
Length=747
Score = 35.4 bits (80), Expect = 0.091, Method: Compositional matrix adjust.
Identities = 24/105 (22%), Positives = 48/105 (45%), Gaps = 7/105 (6%)
Query 35 KVVCLGCVEFFAHLVNQRIAHELLALQLCELLLQQ-----PTNDSVEVCIGFLKAVGQVL 89
K+ LG + L+ Q++ E + + + LL P VE F +G+ L
Sbjct 297 KLRTLGNIRLIGELLKQKMVPEKIVHHIVQELLGDDTKACPAEGDVEALCQFFITIGKQL 356
Query 90 EDV--CRGGFDAAFERLRAILQEGETDKKTQYSIEALWDIRRNGF 132
+D RG D F RL+ + + + + + ++ ++ + D+R N +
Sbjct 357 DDSPRSRGINDTYFGRLKELARHPQLELRLRFMVQNVVDLRANKW 401
> CE25738
Length=1185
Score = 33.5 bits (75), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 38/166 (22%), Positives = 70/166 (42%), Gaps = 29/166 (17%)
Query 18 VIKRLILQ-FRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLALQLCELLLQQPTNDSVE 76
VIK++I+ F ++ + GC+ F N R+ E++ + L+ P+ +++
Sbjct 368 VIKQIIMMTFMELFQNINPKTVGGCLRVFLENKNSRVREEVINIYTASLMTISPSKFNLQ 427
Query 77 VCIGFLKAVGQVLEDVCRGGFDAAFERLR--AILQEGET------------DKKTQYSIE 122
+ L + DV + AAFE+L A L G+T D+ ++ E
Sbjct 428 PLVNILVP---MFHDVKKRVRLAAFEQLSVLAYLLNGKTEIIMKPVRDFEQDQNSRGLTE 484
Query 123 A---------LWDIRRNGFKDF--PGVIEDLDLVEVEDKITHEVDI 157
A L IR +G ++ P +++ DL E E + D+
Sbjct 485 AVTARIRRQVLPRIRYDGLIEYSTPPMMDSFDLAEAEMSLPSNADL 530
> 7297773
Length=188
Score = 32.3 bits (72), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 20/31 (64%), Gaps = 2/31 (6%)
Query 15 GELVIKRLILQFRRAYRRDDKVVCLGCVEFF 45
GELV++ L F R +RD +VCLGC +F
Sbjct 25 GELVLEEL--PFARGPKRDSGIVCLGCYQFL 53
> Hs14211895
Length=1320
Score = 31.6 bits (70), Expect = 1.5, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 25/45 (55%), Gaps = 5/45 (11%)
Query 177 DEYEEDEKKWSALSKEILGEDSTDSD-----ASSVDSEAETSSEE 216
+ +EED ++S + I G DST ++ SS DSE + + EE
Sbjct 330 EAFEEDTGRYSCFASNIYGTDSTSAEIYIEGVSSSDSEGDPNKEE 374
> Hs5174665
Length=596
Score = 31.2 bits (69), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 27/115 (23%), Positives = 50/115 (43%), Gaps = 25/115 (21%)
Query 92 VCRGGFDAAFERLRAILQEGETDKKTQYSIEAL------WDIRRNGFKDFPGVIEDLDLV 145
+ R F A + + ++ G T+ S+EA WD+ +FP + LV
Sbjct 473 IPRSMFGVAVHKGKIVIAGGVTEDGLSASVEAFDLTTNKWDV----MTEFPQERSSISLV 528
Query 146 EVEDKITHEVDIMDPSIKGEEMLNI----FKPQDPDE---YEEDEKKWSALSKEI 193
+ + +I G M+ + F P + ++ YE+D+K+W+ + KEI
Sbjct 529 SLAGSLY--------AIGGFAMIQLESKEFAPTEVNDIWKYEDDKKEWAGMLKEI 575
> CE28476
Length=959
Score = 30.4 bits (67), Expect = 3.4, Method: Composition-based stats.
Identities = 12/37 (32%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 139 IEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQD 175
+++ L E+ KI HE+DI +P E++ ++ QD
Sbjct 228 LQETSLEEIFAKIIHEMDIQEPEFTSEQVRSVLFTQD 264
> YBL034c
Length=1513
Score = 29.6 bits (65), Expect = 4.8, Method: Composition-based stats.
Identities = 18/62 (29%), Positives = 33/62 (53%), Gaps = 5/62 (8%)
Query 135 FPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLN-----IFKPQDPDEYEEDEKKWSAL 189
F ++ D D+V + KI HE++ D + K E++++ F P D + E DE+ A+
Sbjct 963 FNKLLADFDIVSTKMKICHELEKKDANFKVEDIISRESSVSFTPIDNKKSEGDEESDDAV 1022
Query 190 SK 191
+
Sbjct 1023 DE 1024
> At3g22830
Length=406
Score = 28.9 bits (63), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 32/60 (53%), Gaps = 0/60 (0%)
Query 137 GVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYEEDEKKWSALSKEILGE 196
G + + ++ E++ H + D S EE+LN+ K D +E E+ ++ + + EI GE
Sbjct 308 GNMSEFEMSELDKLAMHIQGLGDNSSAREEVLNVEKGNDEEEVEDQQQGYHKENNEIYGE 367
Lambda K H
0.316 0.135 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4255059914
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40