bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0310_orf1
Length=164
Score E
Sequences producing significant alignments: (Bits) Value
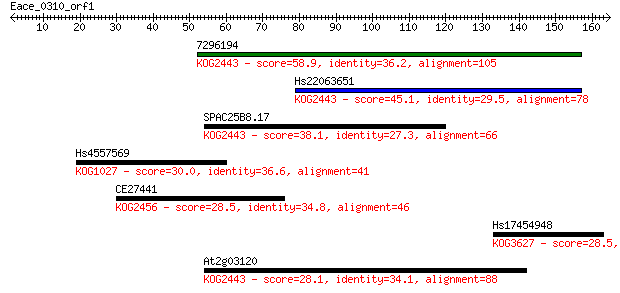
7296194 58.9 5e-09
Hs22063651 45.1 7e-05
SPAC25B8.17 38.1 0.009
Hs4557569 30.0 2.1
CE27441 28.5 6.1
Hs17454948 28.5 6.7
At2g03120 28.1 7.4
> 7296194
Length=389
Score = 58.9 bits (141), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 38/105 (36%), Positives = 57/105 (54%), Gaps = 4/105 (3%)
Query 52 PILYVGSHLSLKQNEVDATTGEKLNKGEAMNRTDAMLFPVFGSIALFSLYLAYKFLGTVW 111
PI++ GS S+K +++ +TGEK + M + DAM FP+ S ALF LYL +K V
Sbjct 54 PIIF-GSIRSVKLHKLKKSTGEK---ADTMTKKDAMYFPLIASAALFGLYLFFKIFQKVH 109
Query 112 VNMLLTLYLTGVGLLALGETTFAVAKPLCPPHLVDDRWLVLHPRG 156
+N LLT Y +G++AL V L P + + +L +G
Sbjct 110 INYLLTGYFFVLGVIALAHLLSPVINSLMPAAVPKVPFHILFTKG 154
> Hs22063651
Length=377
Score = 45.1 bits (105), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 38/78 (48%), Gaps = 0/78 (0%)
Query 79 EAMNRTDAMLFPVFGSIALFSLYLAYKFLGTVWVNMLLTLYLTGVGLLALGETTFAVAKP 138
E + DA FP+ S L LYL +K ++N+LL++Y +G+LAL T
Sbjct 68 ETITSRDAARFPIIASCTLLGLYLFFKIFSQEYINLLLSMYFFVLGILALSHTISPFMNK 127
Query 139 LCPPHLVDDRWLVLHPRG 156
P + ++ +L +G
Sbjct 128 FFPASFPNRQYQLLFTQG 145
> SPAC25B8.17
Length=295
Score = 38.1 bits (87), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 18/66 (27%), Positives = 38/66 (57%), Gaps = 8/66 (12%)
Query 54 LYVGSHLSLKQNEVDATTGEKLNKGEAMNRTDAMLFPVFGSIALFSLYLAYKFLGTVWVN 113
+Y+G+ S ++ E + + + +N+ A+LFP+FG + L +YLA ++L ++
Sbjct 14 VYIGAKWSAQEEEPE--------EKQLINKRLAVLFPIFGGVTLVLMYLALRYLSKEYIQ 65
Query 114 MLLTLY 119
++L Y
Sbjct 66 LILQGY 71
> Hs4557569
Length=977
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 19/41 (46%), Gaps = 0/41 (0%)
Query 19 TLFYSCFVCLAVVMVTPWLYPLPVLLQMGLYTAPILYVGSH 59
TL Y F+ V +T W YP P + P LYVG +
Sbjct 249 TLRYLTFMSGEVGRITKWKYPFPKETEAKSKLTPTLYVGKY 289
> CE27441
Length=493
Score = 28.5 bits (62), Expect = 6.1, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 29/49 (59%), Gaps = 6/49 (12%)
Query 30 VVMVTPWLYPLPVLLQMGLYTAPILYVGSHLSLKQNEVD---ATTGEKL 75
V++++PW YP+ ++L L P + G+ + +K +E+ A T EKL
Sbjct 107 VLIISPWNYPVSMIL---LPMVPAIAAGNTVVIKPSELSENVAATFEKL 152
> Hs17454948
Length=250
Score = 28.5 bits (62), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 19/32 (59%), Gaps = 2/32 (6%)
Query 133 FAVAKPLCPPHLVDDRWLVL--HPRGPFLWLR 162
F + + C L+ DRWL+ H R P+LW+R
Sbjct 41 FHLTRLFCGATLISDRWLLTAAHCRKPYLWVR 72
> At2g03120
Length=344
Score = 28.1 bits (61), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 30/88 (34%), Positives = 42/88 (47%), Gaps = 10/88 (11%)
Query 54 LYVGSHLSLKQNEVDATTGEKLNKGEAMNRTDAMLFPVFGSIALFSLYLAYKFLGTVWVN 113
+YVG S+K E M++ AM FP+ GS L SL+L +KFL VN
Sbjct 38 VYVGCFRSVKDTP----------PTETMSKEHAMRFPLVGSAMLLSLFLLFKFLSKDLVN 87
Query 114 MLLTLYLTGVGLLALGETTFAVAKPLCP 141
+LT Y +G++AL T + P
Sbjct 88 AVLTAYFFVLGIVALSATLLPAIRRFLP 115
Lambda K H
0.325 0.138 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2317343104
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40