bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0298_orf1
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
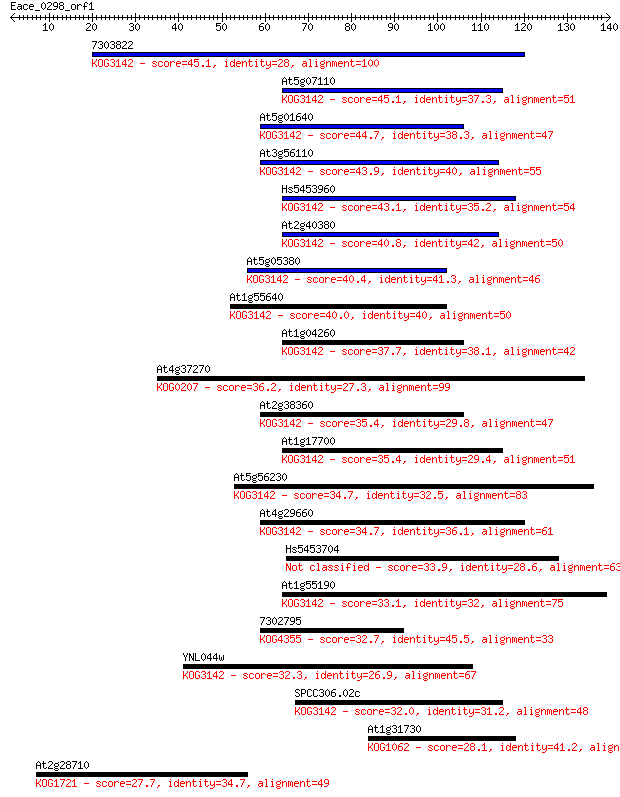
7303822 45.1 4e-05
At5g07110 45.1 5e-05
At5g01640 44.7 5e-05
At3g56110 43.9 1e-04
Hs5453960 43.1 1e-04
At2g40380 40.8 8e-04
At5g05380 40.4 0.001
At1g55640 40.0 0.002
At1g04260 37.7 0.007
At4g37270 36.2 0.019
At2g38360 35.4 0.036
At1g17700 35.4 0.038
At5g56230 34.7 0.054
At4g29660 34.7 0.057
Hs5453704 33.9 0.089
At1g55190 33.1 0.17
7302795 32.7 0.24
YNL044w 32.3 0.28
SPCC306.02c 32.0 0.41
At1g31730 28.1 5.1
At2g28710 27.7 7.7
> 7303822
Length=193
Score = 45.1 bits (105), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 45/100 (45%), Gaps = 6/100 (6%)
Query 20 SPPSPSELSGTLDSHKVPGGAKFAVSQVHGVYLALHRLVLSRCAPWPEFFASSSFQRPGT 79
PP PS ++D +P + L + ++V + PW FF ++F+ +
Sbjct 13 QPPPPSGGRFSVDMQSLPSLSNLPSP------LQIFQMVRNSLRPWVVFFNINNFKTAIS 66
Query 80 GAAAVDRLERNLRYFSTNYICICCVLSLACALLNPALLGV 119
R+ RNL YF NY+ I VL + C + P +L V
Sbjct 67 MQRLNSRVIRNLSYFQANYVFIFFVLMIYCLITAPCILLV 106
> At5g07110
Length=216
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 64 PWPEFFASSSFQRPGTGAAAVDRLERNLRYFSTNYICICCVLSLACALLNP 114
PW E S+F RP + + A R+ +N YF NYI + +L A L +P
Sbjct 46 PWAELVDRSAFSRPPSLSEATSRVRKNFSYFRANYITLVAILLAASLLTHP 96
> At5g01640
Length=223
Score = 44.7 bits (104), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 59 LSRCAPWPEFFASSSFQRPGTGAAAVDRLERNLRYFSTNYICICCVL 105
LSR PW E S+F +P + + A R +N YF NY+CI ++
Sbjct 51 LSRSRPWSELLDRSAFTKPDSLSEAGTRFRKNSSYFRVNYVCIVALI 97
> At3g56110
Length=209
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 22/55 (40%), Positives = 30/55 (54%), Gaps = 1/55 (1%)
Query 59 LSRCAPWPEFFASSSFQRPGTGAAAVDRLERNLRYFSTNYICICCVLSLACALLN 113
LS+ PW E SS RP + A+ R+ +NL YF NY+ I L LA +L +
Sbjct 41 LSQRRPWTELIDRSSMARPESLTDALSRIRKNLAYFKVNYVAIVS-LVLAFSLFS 94
> Hs5453960
Length=185
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 26/54 (48%), Gaps = 0/54 (0%)
Query 64 PWPEFFASSSFQRPGTGAAAVDRLERNLRYFSTNYICICCVLSLACALLNPALL 117
PW F F RP RL RN+ Y+ +NY+ + L L C + +P LL
Sbjct 44 PWSTFVDQQRFSRPRNLGELCQRLVRNVEYYQSNYVFVFLGLILYCVVTSPMLL 97
> At2g40380
Length=213
Score = 40.8 bits (94), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 21/50 (42%), Positives = 27/50 (54%), Gaps = 1/50 (2%)
Query 64 PWPEFFASSSFQRPGTGAAAVDRLERNLRYFSTNYICICCVLSLACALLN 113
PW E SSF RP + + R+ +NL YF NY I L LA +LL+
Sbjct 48 PWLELVDRSSFARPDSLTDSFSRIRKNLAYFKVNYSAIVS-LVLAFSLLS 96
> At5g05380
Length=217
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 24/46 (52%), Gaps = 0/46 (0%)
Query 56 RLVLSRCAPWPEFFASSSFQRPGTGAAAVDRLERNLRYFSTNYICI 101
R LS+ PW E S+ RP + A R+ RNL YF NY+ I
Sbjct 41 RQSLSQRRPWLELVDRSAISRPESLTDAYSRIRRNLPYFKVNYVTI 86
> At1g55640
Length=187
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 32/53 (60%), Gaps = 3/53 (5%)
Query 52 LALHRLV--LSRCAPW-PEFFASSSFQRPGTGAAAVDRLERNLRYFSTNYICI 101
L++H L+ +S PW EF A S RP + + AV R++ NL +F+ NY+ +
Sbjct 25 LSVHNLIASVSSYRPWWSEFLAFGSIDRPSSFSPAVSRVKLNLHHFAVNYVLL 77
> At1g04260
Length=182
Score = 37.7 bits (86), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 0/42 (0%)
Query 64 PWPEFFASSSFQRPGTGAAAVDRLERNLRYFSTNYICICCVL 105
PW +F S+F P + A A R+ +NL +F NY I +L
Sbjct 22 PWGDFLDLSAFSFPSSIADATTRVTQNLTHFRINYSIILSIL 63
> At4g37270
Length=819
Score = 36.2 bits (82), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 42/99 (42%), Gaps = 10/99 (10%)
Query 35 KVPGGAKFAVSQVHGVYLALHRLVLSRCAPWPEFFASSSFQRPGTGAAAVDRLERNLRYF 94
+VPGGA+ + G R+++ W + + Q + +L+R L F
Sbjct 313 RVPGGAR----NLDG------RMIVKATKAWNDSTLNKIVQLTEEAHSNKPKLQRWLDEF 362
Query 95 STNYICICCVLSLACALLNPALLGVAGLCGALCSYASFK 133
NY + VLSLA A L P L L A C + ++
Sbjct 363 GENYSKVVVVLSLAIAFLGPFLFKWPFLSTAACRGSVYR 401
> At2g38360
Length=220
Score = 35.4 bits (80), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 24/47 (51%), Gaps = 0/47 (0%)
Query 59 LSRCAPWPEFFASSSFQRPGTGAAAVDRLERNLRYFSTNYICICCVL 105
LS+ PW E S+ +P + + A R+ +N YF NY+ + +
Sbjct 51 LSKRRPWAELADRSALSKPESISDAAVRIRKNYSYFKVNYLTVATAI 97
> At1g17700
Length=180
Score = 35.4 bits (80), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 20/51 (39%), Gaps = 0/51 (0%)
Query 64 PWPEFFASSSFQRPGTGAAAVDRLERNLRYFSTNYICICCVLSLACALLNP 114
PW + SF P A + R+ N YF TNY + + NP
Sbjct 38 PWKQMLDLGSFNFPRKLATVITRIRANTVYFQTNYTIVVLFSVFLSLIWNP 88
> At5g56230
Length=186
Score = 34.7 bits (78), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 27/88 (30%), Positives = 37/88 (42%), Gaps = 7/88 (7%)
Query 53 ALHRLV--LSRCAPWPEFFASSSFQRPGTGAAAVDRLERNLRYFSTNYICI---CCVLSL 107
++H L +S PW E S F P + ++ + R + N YF NY I C +L
Sbjct 24 SIHNLTTAISSHRPWSELIFSGDFSLPESFSSLLLRSKTNFNYFFVNYTIIVSTCAAFAL 83
Query 108 ACALLNPALLGVAGLCGALCSYASFKGE 135
A +P L V G AL F E
Sbjct 84 ITA--SPVALIVVGAIIALWLIFHFFRE 109
> At4g29660
Length=272
Score = 34.7 bits (78), Expect = 0.057, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 29/61 (47%), Gaps = 0/61 (0%)
Query 59 LSRCAPWPEFFASSSFQRPGTGAAAVDRLERNLRYFSTNYICICCVLSLACALLNPALLG 118
LS PW E S+F P + + A +R++ N+ F TNYI I V L P L
Sbjct 113 LSSQRPWLELVQCSAFSLPISFSVATERIKSNIMIFRTNYIVIFIVSIFISMLWQPVHLS 172
Query 119 V 119
V
Sbjct 173 V 173
> Hs5453704
Length=188
Score = 33.9 bits (76), Expect = 0.089, Method: Compositional matrix adjust.
Identities = 18/66 (27%), Positives = 31/66 (46%), Gaps = 3/66 (4%)
Query 65 WPEFF-ASSSFQRPGTGAAAV--DRLERNLRYFSTNYICICCVLSLACALLNPALLGVAG 121
W +FF S F RP + +R+ NL Y+ TNY+ + ++ L+P + + G
Sbjct 11 WDDFFPGSDRFARPDFRDISKWNNRVVSNLLYYQTNYLVVAAMMISIVGFLSPFNMILGG 70
Query 122 LCGALC 127
+ L
Sbjct 71 IVVVLV 76
> At1g55190
Length=189
Score = 33.1 bits (74), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 34/77 (44%), Gaps = 3/77 (3%)
Query 64 PWPEFFASSSFQRPGTGAAAVDRLERNLRYFSTNY-ICICCVLSLACALLNPALLGVAGL 122
PW F S P A+ R++ NL YF NY I + +L L+ L +P L V +
Sbjct 38 PWKSMFDFESMTLPHGFFDAISRIKTNLGYFRANYAIGVLFILFLSL-LYHPTSLIVLSI 96
Query 123 CGALCSYASF-KGEVLV 138
+ F + E LV
Sbjct 97 LVVFWIFLYFLRDEPLV 113
> 7302795
Length=552
Score = 32.7 bits (73), Expect = 0.24, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 21/35 (60%), Gaps = 2/35 (5%)
Query 59 LSRCAPW--PEFFASSSFQRPGTGAAAVDRLERNL 91
++ CA + P F + F RPGT AA +DR+ NL
Sbjct 385 MTLCAKYRFPSLFINQFFPRPGTPAAKMDRIPANL 419
> YNL044w
Length=176
Score = 32.3 bits (72), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 18/67 (26%), Positives = 30/67 (44%), Gaps = 1/67 (1%)
Query 41 KFAVSQVHGVYLALHRLVLSRCAPWPEFFASSSFQRPGTGAAAVDRLERNLRYFSTNYIC 100
F++ + + +L + + P EFF +P R+ NL+YFS+NY
Sbjct 16 NFSMENIKSEFQSLQSKLATLRTP-QEFFNFKKISKPQNFGEVQSRVAYNLKYFSSNYGL 74
Query 101 ICCVLSL 107
I LS+
Sbjct 75 IIGCLSI 81
> SPCC306.02c
Length=171
Score = 32.0 bits (71), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 22/48 (45%), Gaps = 0/48 (0%)
Query 67 EFFASSSFQRPGTGAAAVDRLERNLRYFSTNYICICCVLSLACALLNP 114
EF RP + A R+ N FS+NY+ I +L + + NP
Sbjct 34 EFLDVRRISRPRNFSEAQSRISFNFSRFSSNYLAIIAMLVIYALIRNP 81
> At1g31730
Length=933
Score = 28.1 bits (61), Expect = 5.1, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 24/38 (63%), Gaps = 6/38 (15%)
Query 84 VDRLERNLRYFSTNYICICCVLSLACALLN----PALL 117
V+ ++++LR S NY+ +C L+ C L+N PA+L
Sbjct 124 VNTIQKDLR--SDNYLVVCAALNAICRLINEETIPAVL 159
> At2g28710
Length=156
Score = 27.7 bits (60), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 26/50 (52%), Gaps = 3/50 (6%)
Query 7 QAAQTRATVCLGGSPPSPSELSGTLDSHKVPG-GAKFAVSQVHGVYLALH 55
+A+ R+ G +PPSP + H+ P GA+FAV Q G ++ H
Sbjct 53 RASHRRSAALEGHAPPSPKRVKPV--KHECPICGAEFAVGQALGGHMRKH 100
Lambda K H
0.321 0.134 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1534984332
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40