bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0217_orf1
Length=253
Score E
Sequences producing significant alignments: (Bits) Value
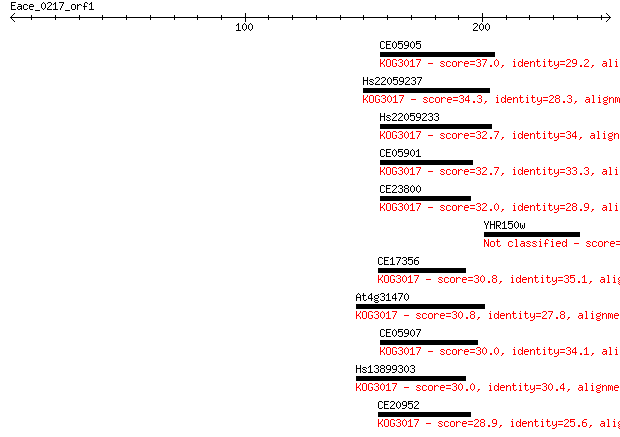
CE05905 37.0 0.040
Hs22059237 34.3 0.23
Hs22059233 32.7 0.77
CE05901 32.7 0.77
CE23800 32.0 1.1
YHR150w 31.2 2.0
CE17356 30.8 2.6
At4g31470 30.8 3.0
CE05907 30.0 4.2
Hs13899303 30.0 5.0
CE20952 28.9 9.4
> CE05905
Length=207
Score = 37.0 bits (84), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 23/48 (47%), Gaps = 0/48 (0%)
Query 157 LLWSQSTKIACAVGVCTDDTPLSSGKEAILVCQFSPAAQQNAAPFSKE 204
+ W+ ++ I C V C D + + + +VCQ+SP P KE
Sbjct 139 MAWANTSSIGCGVKNCGRDASMRNMNKIAVVCQYSPPGNTMGRPIYKE 186
> Hs22059237
Length=344
Score = 34.3 bits (77), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 24/53 (45%), Gaps = 2/53 (3%)
Query 150 DGANLANLLWSQSTKIACAVGVCTDDTPLSSGKEAILVCQFSPAAQQNAAPFS 202
D +N L+W S K+ CAV C+ + AI +C ++P P+
Sbjct 154 DCSNYIQLVWDHSYKVGCAVTPCSKIGHIIHA--AIFICNYAPGGTLTRRPYE 204
> Hs22059233
Length=233
Score = 32.7 bits (73), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 24/48 (50%), Gaps = 4/48 (8%)
Query 157 LLWSQSTKIACAVGVCTDDTPLSSGKEAILVCQFSPAAQ-QNAAPFSK 203
L+W+ S + CAV +C + L AI VC + PA N P+ +
Sbjct 141 LVWANSFYVGCAVAMCPN---LGGASTAIFVCNYGPAGNFANMPPYVR 185
> CE05901
Length=212
Score = 32.7 bits (73), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 13/39 (33%), Positives = 22/39 (56%), Gaps = 3/39 (7%)
Query 157 LLWSQSTKIACAVGVCTDDTPLSSGKEAILVCQFSPAAQ 195
+ W+ + KI C + C+ D S G + ++VC +SPA
Sbjct 147 MAWATTNKIGCGISKCSSD---SFGTQYVVVCLYSPAGN 182
> CE23800
Length=208
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 11/38 (28%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 157 LLWSQSTKIACAVGVCTDDTPLSSGKEAILVCQFSPAA 194
+ W+++ KI C V C D+ +++ + +VCQ+ A
Sbjct 140 MAWAETNKIGCGVKNCGKDSSMNNMYKVAVVCQYDQAG 177
> YHR150w
Length=579
Score = 31.2 bits (69), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 201 FSKEYYDALHARNTPITEMTEADLKESSTGGAAAAVPSLL 240
FSK + D L +RN I E++E D + +TG + L+
Sbjct 315 FSKSFMDTLESRNQEIDEISELDKRTENTGELKQGMKVLI 354
> CE17356
Length=241
Score = 30.8 bits (68), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 13/39 (33%), Positives = 20/39 (51%), Gaps = 2/39 (5%)
Query 156 NLLWSQSTKIACAVGVC--TDDTPLSSGKEAILVCQFSP 192
+LW ++ K+ CAV C D L K + VC++ P
Sbjct 155 QILWKETRKLGCAVQECPARQDGSLDGQKYNVAVCKYYP 193
> At4g31470
Length=185
Score = 30.8 bits (68), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 15/55 (27%), Positives = 25/55 (45%), Gaps = 9/55 (16%)
Query 147 SNPDGANLANLLWSQSTKIACAVGVC-TDDTPLSSGKEAILVCQFSPAAQQNAAP 200
+N D + L+W +S++I CA+ C T DT ++C + P P
Sbjct 137 ANGDCLHYTQLVWKKSSRIGCAISFCKTGDT--------FIICNYDPPGNIVGQP 183
> CE05907
Length=211
Score = 30.0 bits (66), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 22/41 (53%), Gaps = 2/41 (4%)
Query 157 LLWSQSTKIACAVGVCTDDTPLSSGKEAILVCQFSPAAQQN 197
+ W+ + I C V C D L++ A++VCQ+ AQ N
Sbjct 143 MAWANTGLIGCGVKNCGPDPELNNYNRAVVVCQYK--AQGN 181
> Hs13899303
Length=500
Score = 30.0 bits (66), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 23/48 (47%), Gaps = 2/48 (4%)
Query 147 SNPDGANLANLLWSQSTKIACAVGVCTDDTPLSS--GKEAILVCQFSP 192
S P + ++W+ S +I CA+ +C + K LVC +SP
Sbjct 161 SGPVCTHYTQVVWATSNRIGCAINLCHNMNIWGQIWPKAVYLVCNYSP 208
> CE20952
Length=208
Score = 28.9 bits (63), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 10/39 (25%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 156 NLLWSQSTKIACAVGVCTDDTPLSSGKEAILVCQFSPAA 194
+ W++++KI C + C D + + +VCQ+ A
Sbjct 139 QMAWAETSKIGCGIKNCGKDANKKNMYKVAVVCQYDSAG 177
Lambda K H
0.316 0.127 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5223052002
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40