bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
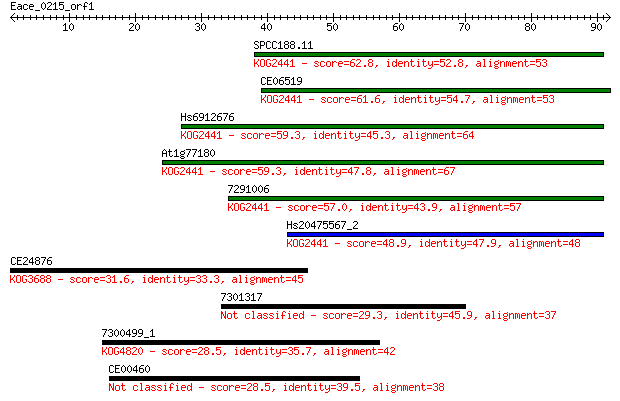
Query= Eace_0215_orf1
Length=91
Score E
Sequences producing significant alignments: (Bits) Value
SPCC188.11 62.8 1e-10
CE06519 61.6 4e-10
Hs6912676 59.3 1e-09
At1g77180 59.3 2e-09
7291006 57.0 9e-09
Hs20475567_2 48.9 2e-06
CE24876 31.6 0.36
7301317 29.3 1.7
7300499_1 28.5 2.8
CE00460 28.5 3.5
> SPCC188.11
Length=557
Score = 62.8 bits (151), Expect = 1e-10, Method: Composition-based stats.
Identities = 28/53 (52%), Positives = 37/53 (69%), Gaps = 0/53 (0%)
Query 38 IPPYLHRQGFVPKCEEDFGGGGSYPEIFVAQFPLGMGKNMDGGLSSSTLSLQV 90
IPPY R+G+ P EDFG GG++PEI VAQ+PL MG+ + +TL+LQV
Sbjct 43 IPPYGQRKGWFPSSPEDFGDGGAFPEIHVAQYPLDMGRKRSAKSAGNTLALQV 95
> CE06519
Length=535
Score = 61.6 bits (148), Expect = 4e-10, Method: Composition-based stats.
Identities = 29/53 (54%), Positives = 35/53 (66%), Gaps = 0/53 (0%)
Query 39 PPYLHRQGFVPKCEEDFGGGGSYPEIFVAQFPLGMGKNMDGGLSSSTLSLQVG 91
PPY R F P+ EDFG GG++PEI VAQFPLG+G G +TL+LQ G
Sbjct 45 PPYGKRTSFRPRGPEDFGDGGAFPEIHVAQFPLGLGLGDMRGKPENTLALQYG 97
> Hs6912676
Length=536
Score = 59.3 bits (142), Expect = 1e-09, Method: Composition-based stats.
Identities = 29/64 (45%), Positives = 42/64 (65%), Gaps = 3/64 (4%)
Query 27 KSTSNNKSSISIPPYLHRQGFVPKCEEDFGGGGSYPEIFVAQFPLGMGKNMDGGLSSSTL 86
+ TS S PPY +R+G++P+ EDFG GG++PEI VAQ+PL MG+ S+ L
Sbjct 30 RQTSLVSSRREPPPYGYRKGWIPRLLEDFGDGGAFPEIHVAQYPLDMGRKKK---MSNAL 86
Query 87 SLQV 90
++QV
Sbjct 87 AIQV 90
> At1g77180
Length=613
Score = 59.3 bits (142), Expect = 2e-09, Method: Composition-based stats.
Identities = 32/70 (45%), Positives = 42/70 (60%), Gaps = 3/70 (4%)
Query 24 RPLKSTSNNKSSIS---IPPYLHRQGFVPKCEEDFGGGGSYPEIFVAQFPLGMGKNMDGG 80
R +S + SSI +P YL+RQG PK EDFG GG++PEI + Q+PL MGKN
Sbjct 29 RVTESETVKSSSIKFKVVPAYLNRQGLRPKNPEDFGDGGAFPEIHLPQYPLLMGKNKSNK 88
Query 81 LSSSTLSLQV 90
+ TL + V
Sbjct 89 PGAKTLPVTV 98
> 7291006
Length=360
Score = 57.0 bits (136), Expect = 9e-09, Method: Composition-based stats.
Identities = 25/57 (43%), Positives = 39/57 (68%), Gaps = 0/57 (0%)
Query 34 SSISIPPYLHRQGFVPKCEEDFGGGGSYPEIFVAQFPLGMGKNMDGGLSSSTLSLQV 90
+ I+ PPY R+ +VP + DFG GG++PEI VAQ+PLG+G + G S L++++
Sbjct 37 AKIAAPPYGQRKDWVPHTDADFGDGGAFPEIHVAQYPLGLGAPGNVGKKSDALAVRL 93
> Hs20475567_2
Length=320
Score = 48.9 bits (115), Expect = 2e-06, Method: Composition-based stats.
Identities = 23/48 (47%), Positives = 33/48 (68%), Gaps = 3/48 (6%)
Query 43 HRQGFVPKCEEDFGGGGSYPEIFVAQFPLGMGKNMDGGLSSSTLSLQV 90
+R+G+ + EDFG GG++PEI VAQ+PL MG+ SS L++QV
Sbjct 3 YRKGWTLRLLEDFGDGGAFPEIHVAQYPLDMGRKKK---ISSALAIQV 47
> CE24876
Length=562
Score = 31.6 bits (70), Expect = 0.36, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 21/45 (46%), Gaps = 0/45 (0%)
Query 1 PRASRLCVSDASLGSYSLSTAPIRPLKSTSNNKSSISIPPYLHRQ 45
P +R+ +D G Y LS P+R T S+ PP+ H Q
Sbjct 133 PYHNRIHAADVLHGCYYLSAHPVRSTFLTPKTPDSVLTPPHPHHQ 177
> 7301317
Length=842
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 25/41 (60%), Gaps = 4/41 (9%)
Query 33 KSSISIPPYLHRQGFVPKCEEDFGGGGS----YPEIFVAQF 69
+S++SI P+L R+G + K EE+ G GS PE A+F
Sbjct 403 RSALSIFPFLLRRGDLRKKEENTMGTGSKNYQAPEWLTAEF 443
> 7300499_1
Length=1558
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 21/42 (50%), Gaps = 2/42 (4%)
Query 15 SYSLSTAPIRPLKSTSNNKSSISIPPYLHRQGFVPKCEEDFG 56
S SL+ AP P K + K+ ++ PP R P E+D G
Sbjct 655 SQSLAKAPKTPCKESKETKADMANPP--RRSSISPVVEDDSG 694
> CE00460
Length=632
Score = 28.5 bits (62), Expect = 3.5, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 5/40 (12%)
Query 16 YSLSTA--PIRPLKSTSNNKSSISIPPYLHRQGFVPKCEE 53
YS TA P PLK + S + +P Y+H VP C++
Sbjct 346 YSEETAKIPNNPLKGSV---SDVKLPEYVHEIKMVPHCDD 382
Lambda K H
0.314 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174483934
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40