bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0214_orf1
Length=173
Score E
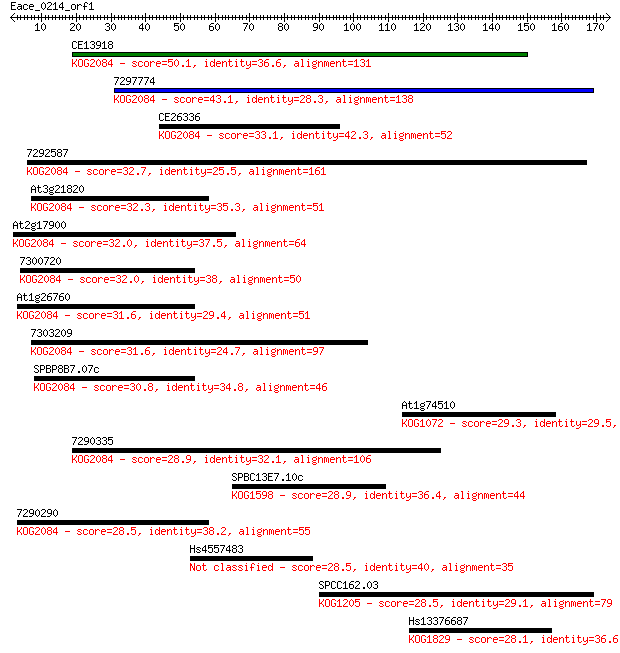
Sequences producing significant alignments: (Bits) Value
CE13918 50.1 2e-06
7297774 43.1 3e-04
CE26336 33.1 0.32
7292587 32.7 0.35
At3g21820 32.3 0.42
At2g17900 32.0 0.62
7300720 32.0 0.67
At1g26760 31.6 0.76
7303209 31.6 0.84
SPBP8B7.07c 30.8 1.3
At1g74510 29.3 3.9
7290335 28.9 5.0
SPBC13E7.10c 28.9 5.5
7290290 28.5 7.3
Hs4557483 28.5 7.6
SPCC162.03 28.5 7.6
Hs13376687 28.1 9.9
> CE13918
Length=501
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 48/140 (34%), Positives = 67/140 (47%), Gaps = 20/140 (14%)
Query 19 EITISYIGDEDLLKSTNIRRDKL-SGWLFTCYCTRCIDTSDPA-RGFRC--PACGTGTMQ 74
E ISYI D+ +S IRR L S W F C CTRC+D D A RC PAC +
Sbjct 240 EAFISYI---DVGRSKYIRRRDLNSRWYFNCECTRCMDPEDDALTAIRCANPACDAPILT 296
Query 75 FKTEQDETTANPCTVCGTVADASTV---ERYL-TFEAAYAERLD-ETNREDMKDAEMVYE 129
+TE+ A C C T+ + TV + Y+ T A++ + E +E + AE +
Sbjct 297 SETEEPMNIA--CEKCKTIVEEDTVKAAQEYMKTLPASFDPKCPAEILKELLAKAEQIL- 353
Query 130 QASRVFTSHWILFQLNTILF 149
S V+ + +L T LF
Sbjct 354 HPSNVYVA-----RLRTALF 368
> 7297774
Length=507
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 39/147 (26%), Positives = 60/147 (40%), Gaps = 9/147 (6%)
Query 31 LKSTNIRRDKLS-GWLFTCYCTRCIDTSDPARGFRCPACGTGTMQFKTEQDET---TANP 86
L T R+ L G FTC C RC+D ++ F CG F+ + T T+
Sbjct 256 LDGTAQRQKHLKQGKFFTCQCERCLDPTELGTHFSSLKCGQCAEGFQVPRQPTEPDTSWN 315
Query 87 CTVCG---TVADASTVERYLTFEAAYAERLDETNREDMKDAEMVYEQASRVFTSHWILFQ 143
C CG + ADA + + L E + L + + ++ + S + H+I
Sbjct 316 CANCGSDTSNADALAMLQSLQSEVNAVQALPMAAKRLEEIERLLRKYKSLLHPLHFIATG 375
Query 144 LNTILFEGYRAAGDYESAYV--HQMHR 168
L +L E Y YE + HQ+ R
Sbjct 376 LRQLLIEMYGRVQGYEMVQLPDHQLER 402
> CE26336
Length=538
Score = 33.1 bits (74), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 25/62 (40%), Gaps = 10/62 (16%)
Query 44 WLFTCYCTRCIDTSDP-ARGFRCPACGTGTMQFKTEQ---DETTANP------CTVCGTV 93
W F C+C RC D D CP C + T KT + E ANP C C T
Sbjct 235 WYFECHCERCDDPDDNWLTSVLCPVCISKTEYRKTIKLHGPEAYANPETLEIVCDRCSTT 294
Query 94 AD 95
D
Sbjct 295 LD 296
> 7292587
Length=539
Score = 32.7 bits (73), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 41/165 (24%), Positives = 65/165 (39%), Gaps = 12/165 (7%)
Query 6 FVLKARRALPAEGEITISYIGDEDLLKSTNIRRDKL-SGWLFTCYCTRCIDTSDPA---R 61
+L+ R + +T+SY L+ T RR + G LF C C RC D +
Sbjct 281 ILLRTSRRVREREALTLSYAYT---LQGTLKRRAFMHEGKLFWCCCRRCSDPRELGTDCS 337
Query 62 GFRCPACGTGTMQFKTEQDETTANPCTVCGTVADASTVERYLTFEAAYAERLDETNREDM 121
C C TG+++ +T C C A+ VER L E +D + +
Sbjct 338 ALVCATCRTGSVRAVDPLQQTGDWACDRCAHKMGATEVERQLDRINNDLEDIDVHDIPGL 397
Query 122 KDAEMVYEQASRVFTSHWILFQLNTILFEGYRAAGDYESAYVHQM 166
++ + Y R +H++L L + Y G E + QM
Sbjct 398 ENFLLRYRDVLR--PNHYLLLSAKYSLCQIY---GRTEGYLLPQM 437
> At3g21820
Length=460
Score = 32.3 bits (72), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 30/51 (58%), Gaps = 3/51 (5%)
Query 7 VLKARRALPAEGEITISYIGDEDLLKSTNIRRDKLSGWLFTCYCTRCIDTS 57
V+ A R + E+TISYI +E K R+ L+ + F+C C++C++ S
Sbjct 410 VIIALRRISKNEEVTISYIDEELPYKE---RQALLADYGFSCKCSKCLEDS 457
> At2g17900
Length=447
Score = 32.0 bits (71), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 33/73 (45%), Gaps = 12/73 (16%)
Query 2 EADAFVLKARRALPAEGEITISYIGDEDLLKSTNIRRDKLS-GWLFTCYCTRCIDTSDP- 59
E V++A + + EITISYI + ST R+ L +LF C C RC + P
Sbjct 192 EEQMAVVRAMDNISKDSEITISYI---ETAGSTLTRQKSLKEQYLFHCQCARCSNFGKPH 248
Query 60 -------ARGFRC 65
G+RC
Sbjct 249 DIEESAILEGYRC 261
> 7300720
Length=384
Score = 32.0 bits (71), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 28/52 (53%), Gaps = 3/52 (5%)
Query 4 DAFVLKARRALPAEGEITISYIGDEDLLKSTNIRRDKL--SGWLFTCYCTRC 53
D VLKA + EI ISY+ DE +L+ + R K+ ++F C C +C
Sbjct 304 DIVVLKALAPIQQGEEICISYL-DECMLERSRHSRHKVLRENYVFICQCPKC 354
> At1g26760
Length=967
Score = 31.6 bits (70), Expect = 0.76, Method: Composition-based stats.
Identities = 15/51 (29%), Positives = 24/51 (47%), Gaps = 3/51 (5%)
Query 3 ADAFVLKARRALPAEGEITISYIGDEDLLKSTNIRRDKLSGWLFTCYCTRC 53
D ++ A R + EI+ +Y D+L R++ W F C C+RC
Sbjct 776 GDYVIVHASRDIKTGEEISFAYF---DVLSPLEKRKEMAESWGFCCGCSRC 823
> 7303209
Length=513
Score = 31.6 bits (70), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 24/101 (23%), Positives = 41/101 (40%), Gaps = 6/101 (5%)
Query 7 VLKARRALPAEGEITISYIGDEDLLKSTNIRRDKLSGWLFTCYCTRCIDTSDPARGFRCP 66
+L A R + ++I Y + + + + +R + LF C C RC+D ++ +
Sbjct 257 ILWAPREIKKNAHLSICY--SDAMWGTADRQRHLMQTKLFKCACERCVDVTELDTNYSAI 314
Query 67 AC---GTGTMQFKTEQDETTAN-PCTVCGTVADASTVERYL 103
C G + T+ DE + C C VER L
Sbjct 315 KCEDRQCGGLMLPTKADEWNGSWRCRECHKQVQKHYVERIL 355
> SPBP8B7.07c
Length=483
Score = 30.8 bits (68), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 28/47 (59%), Gaps = 4/47 (8%)
Query 8 LKARRALPAEGEITISYIGDEDLLKSTNIRRDKL-SGWLFTCYCTRC 53
L ++R + + ++ ISYI D+ +IR+ +L + F+CYC RC
Sbjct 211 LVSKRDIKKDEQLFISYI---DIRLPKSIRQKQLLKKYFFSCYCPRC 254
> At1g74510
Length=451
Score = 29.3 bits (64), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 13/44 (29%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 114 DETNREDMKDAEMVYEQASRVFTSHWILFQLNTILFEGYRAAGD 157
+ T R +KD+E+ + ++ HWI F + +E Y GD
Sbjct 123 NRTFRSLIKDSELYRLRRAKGIVEHWIYFSCRLLEWEAYDPNGD 166
> 7290335
Length=430
Score = 28.9 bits (63), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 34/113 (30%), Positives = 44/113 (38%), Gaps = 25/113 (22%)
Query 19 EITISYIGDEDLLKSTNIRR-DKLSGWLFTCYCTRCIDTSDPARGFR--CP--ACGTGTM 73
+I ISYI DLL + RR D + F C C++C D + CP CG G
Sbjct 250 KIFISYI---DLLNTPEQRRLDLKEHYYFLCVCSKCTDAKESKEMLAALCPNRNCGAGI- 305
Query 74 QFKTEQDETTANPCTVCGTVADASTVERYLTFEAAYAERLDET--NREDMKDA 124
N C C DA + A+ E + T N E+MKD
Sbjct 306 -------SVDRNNCPRC----DAGISPK---LRNAFNEAMTLTRHNLENMKDV 344
> SPBC13E7.10c
Length=500
Score = 28.9 bits (63), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 21/45 (46%), Gaps = 5/45 (11%)
Query 65 CPACGTGTMQFKTEQDETTANP-CTVCGTVADASTVERYLTFEAA 108
CP CG+ T E D + N CT CG V + + +TF A
Sbjct 11 CPNCGSTTF----ESDTASGNTYCTQCGVVVEQDAIVSEVTFGEA 51
> 7290290
Length=991
Score = 28.5 bits (62), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 27/56 (48%), Gaps = 4/56 (7%)
Query 3 ADAFVLKARRALPAEGEITISYIGDEDLLKSTNIRRDKLS-GWLFTCYCTRCIDTS 57
+ V+ A +PA EIT+SY LL ST R+ L F C C RC D +
Sbjct 251 GETIVVCATERIPAGAEITMSY---AKLLWSTLARKIFLGMTKHFICKCVRCQDPT 303
> Hs4557483
Length=757
Score = 28.5 bits (62), Expect = 7.6, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 17/35 (48%), Gaps = 0/35 (0%)
Query 53 CIDTSDPARGFRCPACGTGTMQFKTEQDETTANPC 87
CI T R CPA TG T+ +E A+PC
Sbjct 102 CIQTESGGRCGPCPAGFTGNGSHCTDVNECNAHPC 136
> SPCC162.03
Length=292
Score = 28.5 bits (62), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 35/79 (44%), Gaps = 12/79 (15%)
Query 90 CGTVADASTVERYLTFEAAYAERLDETNREDMKDAEMVYEQASRVFTSHWILFQLNTILF 149
C D T+E + +LD T D+K E ++ A R F + I+ I
Sbjct 35 CSRAPDTITIEHSKLLKL----KLDVT---DVKSVETAFKDAKRRFGNVDIV-----INN 82
Query 150 EGYRAAGDYESAYVHQMHR 168
GY G++ES + +MHR
Sbjct 83 AGYGLVGEFESYNIEEMHR 101
> Hs13376687
Length=635
Score = 28.1 bits (61), Expect = 9.9, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 116 TNREDMKDAEMVYEQASRVFTSHWILFQLNTILFEGYRAAG 156
T D AE++ ++ RVF WIL +N+ L AAG
Sbjct 331 TYEPDFNSAELLAKELYRVFQKCWILSVVNSQLAGSLSAAG 371
Lambda K H
0.320 0.133 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2598880752
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40