bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0182_orf1
Length=117
Score E
Sequences producing significant alignments: (Bits) Value
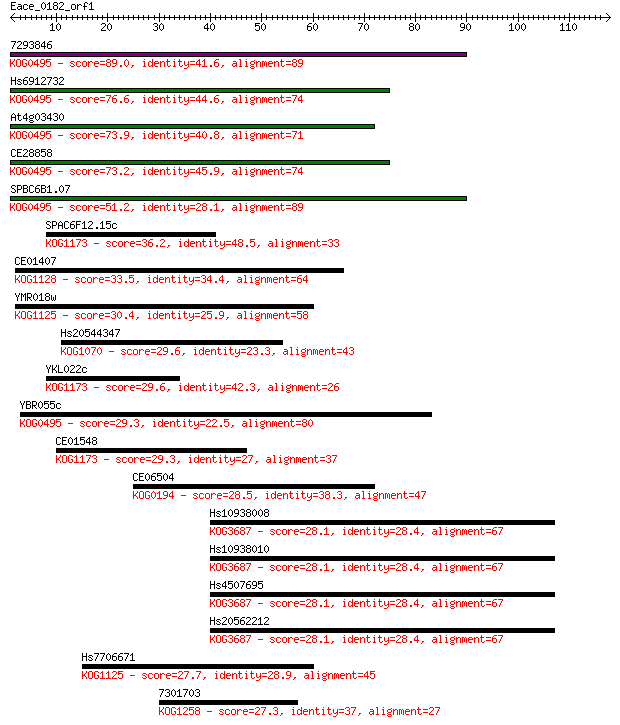
7293846 89.0 2e-18
Hs6912732 76.6 1e-14
At4g03430 73.9 6e-14
CE28858 73.2 1e-13
SPBC6B1.07 51.2 5e-07
SPAC6F12.15c 36.2 0.015
CE01407 33.5 0.093
YMR018w 30.4 0.90
Hs20544347 29.6 1.3
YKL022c 29.6 1.4
YBR055c 29.3 1.7
CE01548 29.3 1.9
CE06504 28.5 2.9
Hs10938008 28.1 4.2
Hs10938010 28.1 4.2
Hs4507695 28.1 4.3
Hs20562212 28.1 4.5
Hs7706671 27.7 5.3
7301703 27.3 6.8
> 7293846
Length=913
Score = 89.0 bits (219), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 37/89 (41%), Positives = 56/89 (62%), Gaps = 0/89 (0%)
Query 1 RLFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNR 60
+LFW + K +K R W NR+V +DP LGDAWA F FE+ HG E + +V++RC A+P
Sbjct 821 KLFWSEHKFSKCRDWFNRTVKIDPDLGDAWAYFYKFELLHGTEAQQQEVLDRCISAEPTH 880
Query 61 GLAWNKTTKQVRCWSFSPAQKLNAVLEYM 89
G +W + +K ++ W F + L AV+ +
Sbjct 881 GESWCRVSKNIQNWQFKTPEVLRAVVREL 909
> Hs6912732
Length=941
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 33/74 (44%), Positives = 47/74 (63%), Gaps = 0/74 (0%)
Query 1 RLFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNR 60
+LFW +KI KAR+W +R+V +D LGDAWA F FE++HG E++ +V RC A+P
Sbjct 849 KLFWSQRKITKAREWFHRTVKIDSDLGDAWAFFYKFELQHGTEEQQEEVRKRCESAEPRH 908
Query 61 GLAWNKTTKQVRCW 74
G W +K + W
Sbjct 909 GELWCAVSKDIANW 922
> At4g03430
Length=1029
Score = 73.9 bits (180), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 29/71 (40%), Positives = 46/71 (64%), Gaps = 0/71 (0%)
Query 1 RLFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNR 60
+LFW+D+K+ KAR W R+VT+ P +GD WA F FE++HG +++ +V+ +C +P
Sbjct 934 KLFWQDKKVEKARAWFERAVTVGPDIGDFWALFYKFELQHGSDEDRKEVVAKCVACEPKH 993
Query 61 GLAWNKTTKQV 71
G W +K V
Sbjct 994 GEKWQAISKAV 1004
> CE28858
Length=968
Score = 73.2 bits (178), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 34/74 (45%), Positives = 46/74 (62%), Gaps = 0/74 (0%)
Query 1 RLFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNR 60
RLFW ++KI KAR+W R+V LDP GDA+A+FLAFE HG E++ V +C ++P
Sbjct 876 RLFWSERKIKKAREWFVRAVNLDPDNGDAFANFLAFEQIHGKEEDRKSVFKKCVTSEPRY 935
Query 61 GLAWNKTTKQVRCW 74
G W +K W
Sbjct 936 GDLWQSVSKDPINW 949
> SPBC6B1.07
Length=906
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 43/89 (48%), Gaps = 0/89 (0%)
Query 1 RLFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNR 60
R+ W ++K KAR W ++V D GD W F + +E G E + +V+ A P+
Sbjct 818 RMLWLEKKADKARSWFLKAVKADQDNGDVWCWFYKYSLEAGNEDQQKEVLTSFETADPHH 877
Query 61 GLAWNKTTKQVRCWSFSPAQKLNAVLEYM 89
G W TK ++ +P + L+ + +
Sbjct 878 GYFWPSITKDIKNSRKTPQELLHLAINVL 906
> SPAC6F12.15c
Length=671
Score = 36.2 bits (82), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 23/36 (63%), Gaps = 3/36 (8%)
Query 8 KIAKARKWMNRSVTLDPSLGDAWASF---LAFEIEH 40
KI++AR++ ++S T+DP G AW F A E EH
Sbjct 388 KISEARRYFSKSSTMDPQFGPAWIGFAHSFAIEGEH 423
> CE01407
Length=787
Score = 33.5 bits (75), Expect = 0.093, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 33/66 (50%), Gaps = 4/66 (6%)
Query 2 LFWKDQKIAKARKWMNRSVTLDPSLGDAW--ASFLAFEIEHGGEKECIDVINRCALAQPN 59
L D+K +A K + RS+ L P W A + A+++E+ KE +RC QP+
Sbjct 476 LLLMDKKFEEAYKHLRRSLELQPIQLGTWFNAGYCAWKLENF--KESTQCYHRCVSLQPD 533
Query 60 RGLAWN 65
AWN
Sbjct 534 HFEAWN 539
> YMR018w
Length=514
Score = 30.4 bits (67), Expect = 0.90, Method: Composition-based stats.
Identities = 15/58 (25%), Positives = 27/58 (46%), Gaps = 0/58 (0%)
Query 2 LFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPN 59
L++ DQKI +++K + + P+ G W + A I+ N+C +PN
Sbjct 369 LYYSDQKIKQSQKCLEFLLLEKPNNGTIWNRYGAILANTKSYHSAINAYNKCKQLRPN 426
> Hs20544347
Length=1871
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 10/43 (23%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 11 KARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRC 53
+A+ +++ P D W+ ++ I+HG +K+ D+ R
Sbjct 1779 RAKAIFENTLSTYPKRTDVWSVYIDMTIKHGSQKDVRDIFERV 1821
> YKL022c
Length=840
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 11/26 (42%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 8 KIAKARKWMNRSVTLDPSLGDAWASF 33
+I++A+K+ ++S LDPS AW F
Sbjct 546 RISEAQKYYSKSSILDPSFAAAWLGF 571
> YBR055c
Length=899
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 18/80 (22%), Positives = 33/80 (41%), Gaps = 2/80 (2%)
Query 3 FWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNRGL 62
F+ + + + KW+ R++ GD W G K+ +D+ N +P G
Sbjct 815 FYAEAQYETSLKWLERALKKCSRYGDTWVWLFRTYARLG--KDTVDLYNMFDQCEPTYGP 872
Query 63 AWNKTTKQVRCWSFSPAQKL 82
W +K V+ +P + L
Sbjct 873 EWIAASKNVKMQYCTPREIL 892
> CE01548
Length=655
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 10/40 (25%), Positives = 22/40 (55%), Gaps = 3/40 (7%)
Query 10 AKARKWMNRSVTLDPSLGDAWASF---LAFEIEHGGEKEC 46
++AR ++++ +D + + W +F L +E+EH C
Sbjct 359 SRARNFISKCTMMDSTFAEGWVAFGHILHYEVEHEQSMSC 398
> CE06504
Length=541
Score = 28.5 bits (62), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 26/51 (50%), Gaps = 4/51 (7%)
Query 25 SLGDAWASFLAFE----IEHGGEKECIDVINRCALAQPNRGLAWNKTTKQV 71
SL DA A L E +E+G KE + +I+ C PN L +N +Q+
Sbjct 381 SLKDAKAWILTEERPHKMENGDPKELMVLIDACCERNPNERLNFNTVKRQI 431
> Hs10938008
Length=1764
Score = 28.1 bits (61), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 31/72 (43%), Gaps = 8/72 (11%)
Query 40 HGGEKECIDVINRCALAQPNRGLAWNKTTKQVRCWSFSPA-----QKLNAVLEYMYPNAL 94
HG ++ +++ RCA +P L R S PA Q L A++E + +
Sbjct 401 HGSQERYFELVERCADQRPESSLL---NLISYRAQSIHPAKDGWIQNLQALMERFFRSES 457
Query 95 KDGISQVVLDAL 106
+ + VLD L
Sbjct 458 RGAVRIKVLDVL 469
> Hs10938010
Length=1763
Score = 28.1 bits (61), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 31/72 (43%), Gaps = 8/72 (11%)
Query 40 HGGEKECIDVINRCALAQPNRGLAWNKTTKQVRCWSFSPA-----QKLNAVLEYMYPNAL 94
HG ++ +++ RCA +P L R S PA Q L A++E + +
Sbjct 401 HGSQERYFELVERCADQRPESSLL---NLISYRAQSIHPAKDGWIQNLQALMERFFRSES 457
Query 95 KDGISQVVLDAL 106
+ + VLD L
Sbjct 458 RGAVRIKVLDVL 469
> Hs4507695
Length=1807
Score = 28.1 bits (61), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 31/72 (43%), Gaps = 8/72 (11%)
Query 40 HGGEKECIDVINRCALAQPNRGLAWNKTTKQVRCWSFSPA-----QKLNAVLEYMYPNAL 94
HG ++ +++ RCA +P L R S PA Q L A++E + +
Sbjct 401 HGSQERYFELVERCADQRPESSLL---NLISYRAQSIHPAKDGWIQNLQALMERFFRSES 457
Query 95 KDGISQVVLDAL 106
+ + VLD L
Sbjct 458 RGAVRIKVLDVL 469
> Hs20562212
Length=1807
Score = 28.1 bits (61), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 31/72 (43%), Gaps = 8/72 (11%)
Query 40 HGGEKECIDVINRCALAQPNRGLAWNKTTKQVRCWSFSPA-----QKLNAVLEYMYPNAL 94
HG ++ +++ RCA +P L R S PA Q L A++E + +
Sbjct 401 HGSQERYFELVERCADQRPESSLL---NLISYRAQSIHPAKDGWIQNLQALMERFFRSES 457
Query 95 KDGISQVVLDAL 106
+ + VLD L
Sbjct 458 RGAVRIKVLDVL 469
> Hs7706671
Length=626
Score = 27.7 bits (60), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 21/45 (46%), Gaps = 0/45 (0%)
Query 15 WMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPN 59
+M ++ DP +AW + E+ E+ I + RC QPN
Sbjct 348 FMEAAILQDPGDAEAWQFLGITQAENENEQAAIVALQRCLELQPN 392
> 7301703
Length=1009
Score = 27.3 bits (59), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 10/27 (37%), Positives = 15/27 (55%), Gaps = 0/27 (0%)
Query 30 WASFLAFEIEHGGEKECIDVINRCALA 56
W +L FEIE G + + + RC +A
Sbjct 651 WKDYLDFEIEKGDRERVLVLFERCLIA 677
Lambda K H
0.319 0.132 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171470346
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40