bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0180_orf1
Length=183
Score E
Sequences producing significant alignments: (Bits) Value
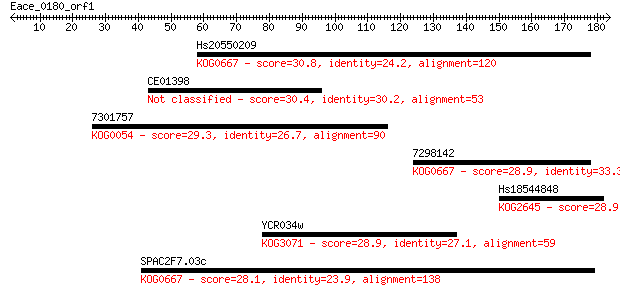
Hs20550209 30.8 1.5
CE01398 30.4 2.3
7301757 29.3 5.0
7298142 28.9 5.7
Hs18544848 28.9 5.9
YCR034w 28.9 6.5
SPAC2F7.03c 28.1 9.4
> Hs20550209
Length=520
Score = 30.8 bits (68), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 29/130 (22%), Positives = 50/130 (38%), Gaps = 10/130 (7%)
Query 58 ALSMAILGGLLYYYSKKPTHLSITLALGMSLAILFMSGFIWFSILLSTHLQLWLADIV-- 115
++ +LG LY K +L++ + + S+ H L +IV
Sbjct 180 CITFELLGINLYELMKNNNFQGFSLSIVRRFTLSVLKCLQMLSVEKIIHCDLKPENIVLY 239
Query 116 TQLEFILYVLDFGASCLALFQLQYVFDSPTYRGPRSFL--------PRWAESAARAEGWK 167
+ + + V+DFG+SC ++ S YR P L W+ AE +
Sbjct 240 QKGQASVKVIDFGSSCYEHQKVYTYIQSRFYRSPEVILGHPYDVAIDMWSLGCITAELYT 299
Query 168 GWSPFQGEGQ 177
G+ F GE +
Sbjct 300 GYPLFPGENE 309
> CE01398
Length=994
Score = 30.4 bits (67), Expect = 2.3, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 30/54 (55%), Gaps = 1/54 (1%)
Query 43 FLSLIAHFFVQDVGGALSMAILGGLLYYYSKKPTHL-SITLALGMSLAILFMSG 95
FL+ F + S+ ++G ++ Y +KKP + ++TL LG+ + + F SG
Sbjct 448 FLAFHGKFTTVKDDPSASLELIGDIIVYLNKKPEQIFNLTLILGLVIIVGFASG 501
> 7301757
Length=1346
Score = 29.3 bits (64), Expect = 5.0, Method: Composition-based stats.
Identities = 24/103 (23%), Positives = 49/103 (47%), Gaps = 13/103 (12%)
Query 26 DEFEKSKPRV----LSIAGYCFLSL-IAHFFVQDVGGALSMAILGGLLYYYSKKPTHLS- 79
DE ++S P V L G F+ + +A + + +L LGGL+ Y++ T +S
Sbjct 40 DELKRSNPSVVRMILRAYGKIFVPMGLAFSISETICKSLMPLFLGGLVGYFATNQTDISE 99
Query 80 -------ITLALGMSLAILFMSGFIWFSILLSTHLQLWLADIV 115
+ + + M + ++ FI++ + T L+L L+ ++
Sbjct 100 NTAYLYAMGIVICMLVPVITFHPFIFYIFQVGTKLRLALSGLI 142
> 7298142
Length=722
Score = 28.9 bits (63), Expect = 5.7, Method: Composition-based stats.
Identities = 18/62 (29%), Positives = 26/62 (41%), Gaps = 8/62 (12%)
Query 124 VLDFGASCLALFQLQYVFDSPTYRGPRSFL--------PRWAESAARAEGWKGWSPFQGE 175
V+DFG+SC ++ S YR P L W+ AE + G+ F GE
Sbjct 342 VIDFGSSCYVDRKIYTYIQSRFYRSPEVILGLQYGTAIDMWSLGCILAELYTGFPLFPGE 401
Query 176 GQ 177
+
Sbjct 402 NE 403
> Hs18544848
Length=440
Score = 28.9 bits (63), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 16/33 (48%), Gaps = 1/33 (3%)
Query 150 RSFLPRWAESAARAEGW-KGWSPFQGEGQRLGG 181
R LP W S R EGW +GW + E + G
Sbjct 333 REMLPFWMNSTGRREGWQRGWHGYDNELMDMRG 365
> YCR034w
Length=347
Score = 28.9 bits (63), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 16/59 (27%), Positives = 32/59 (54%), Gaps = 6/59 (10%)
Query 78 LSITLALGMSLAILFMSGFIWFSILLSTHLQLWLADIVTQLEFILYVLDFGASCLALFQ 136
+ I+L LG+ + + W+ L + +++W + VT+ + I +VLD G A++Q
Sbjct 202 VPISLNLGVHVVMY------WYYFLAARGIRVWWKEWVTRFQIIQFVLDIGFIYFAVYQ 254
> SPAC2F7.03c
Length=1087
Score = 28.1 bits (61), Expect = 9.4, Method: Composition-based stats.
Identities = 33/148 (22%), Positives = 56/148 (37%), Gaps = 11/148 (7%)
Query 41 YCFLSLIAHFFVQDVGGALSMAILGGLLYYYSKKPTHLSITLALGMSLAILFMSGFIWFS 100
YC + HF+ +D ++ +LG LY K + + + S+ + +
Sbjct 759 YCMVQYTDHFYFRD-HLCVATELLGKNLYELIKSNGFKGLPIVVIKSITRQLIQCLTLLN 817
Query 101 ILLSTHLQLWLADIVTQLEFI--LYVLDFGASCLALFQLQYVFDSPTYRGPRSFLPR--- 155
H L +I+ F + V+DFG+SC + S YR P L
Sbjct 818 EKHVIHCDLKPENILLCHPFKSQVKVIDFGSSCFEGECVYTYIQSRFYRSPEVILGMGYG 877
Query 156 -----WAESAARAEGWKGWSPFQGEGQR 178
W+ AE + G+ F GE ++
Sbjct 878 TPIDVWSLGCIIAEMYTGFPLFPGENEQ 905
Lambda K H
0.326 0.142 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2914594326
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40