bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0171_orf1
Length=217
Score E
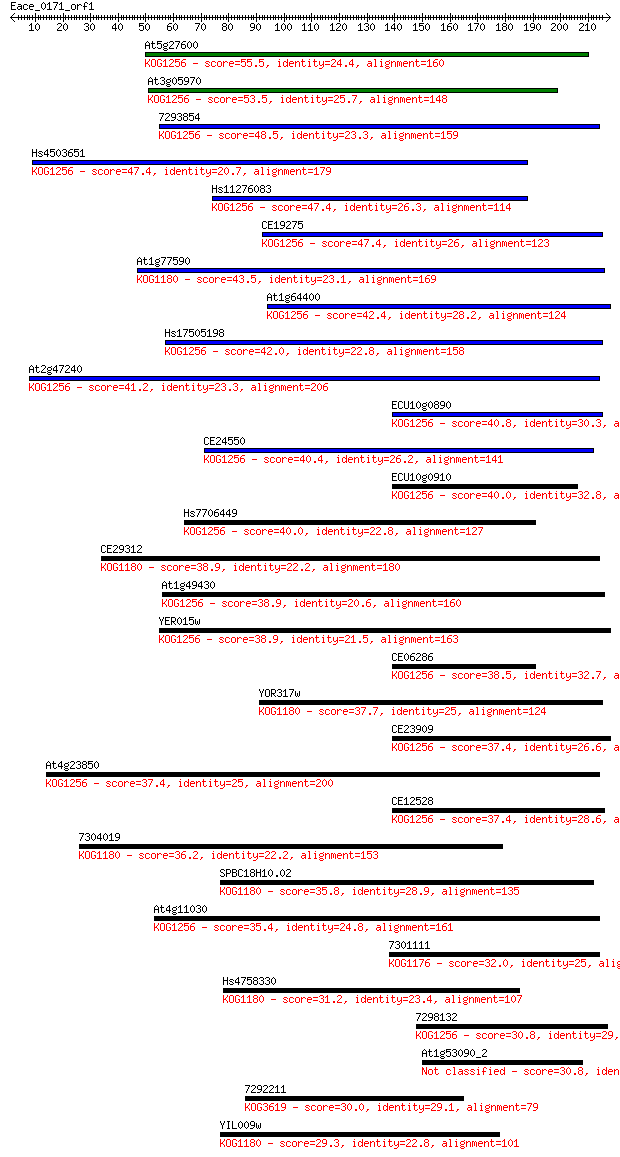
Sequences producing significant alignments: (Bits) Value
At5g27600 55.5 9e-08
At3g05970 53.5 3e-07
7293854 48.5 1e-05
Hs4503651 47.4 2e-05
Hs11276083 47.4 2e-05
CE19275 47.4 2e-05
At1g77590 43.5 4e-04
At1g64400 42.4 6e-04
Hs17505198 42.0 0.001
At2g47240 41.2 0.001
ECU10g0890 40.8 0.002
CE24550 40.4 0.003
ECU10g0910 40.0 0.003
Hs7706449 40.0 0.004
CE29312 38.9 0.007
At1g49430 38.9 0.009
YER015w 38.9 0.009
CE06286 38.5 0.010
YOR317w 37.7 0.018
CE23909 37.4 0.023
At4g23850 37.4 0.025
CE12528 37.4 0.026
7304019 36.2 0.046
SPBC18H10.02 35.8 0.068
At4g11030 35.4 0.077
7301111 32.0 1.0
Hs4758330 31.2 1.6
7298132 30.8 2.1
At1g53090_2 30.8 2.1
7292211 30.0 4.1
YIL009w 29.3 7.2
> At5g27600
Length=698
Score = 55.5 bits (132), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 39/160 (24%), Positives = 65/160 (40%), Gaps = 20/160 (12%)
Query 50 IPQFPHIKSTYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEAL 109
P P I + + A + N Y+G R S T+ ++ + TY + +A+
Sbjct 77 FPDHPEIGTLHDNFVHAVETYAENKYLGTR-----VRSDGTIGEYSWMTYGEAASERQAI 131
Query 110 GRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDT 169
G L L H +G +GL R +W ++D A V+VPLYDT
Sbjct 132 GSGL-------LFHGVNQG--------DCVGLYFINRPEWLVVDHACAAYSFVSVPLYDT 176
Query 170 LGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEGFPLKA 209
LG +VK +V ++ + L+ + G P+ +
Sbjct 177 LGPDAVKFVVNHANLQAIFCVPQTLNIVIAKLPSGNPIHS 216
> At3g05970
Length=657
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 38/148 (25%), Positives = 63/148 (42%), Gaps = 20/148 (13%)
Query 51 PQFPHIKSTYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALG 110
P P I + + + A F Y+G R + T+ D+K+ TY + ALG
Sbjct 78 PDHPDIATLHDNFEHAVHDFRDYKYLGTRVRV-----DGTVGDYKWMTYGEAGTARTALG 132
Query 111 RALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTL 170
L G+P+ +G+ R +W ++D A + V+VPLYDTL
Sbjct 133 SGLVHH-GIPMG--------------SSVGIYFINRPEWLIVDHACSSYSYVSVPLYDTL 177
Query 171 GEPSVKHIVKLTKMEVLAAEGSKLDAAL 198
G +VK IV ++ + L++ +
Sbjct 178 GPDAVKFIVNHATVQAIFCVAETLNSVV 205
> 7293854
Length=704
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 37/159 (23%), Positives = 69/159 (43%), Gaps = 24/159 (15%)
Query 55 HIKSTYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALD 114
++++ Y +E A +G R+ T S +++ Y + L + G +
Sbjct 100 NVRTLYQTFREGAYASNNGPCLGWRE--------TLTSPYQWINYDEALLRAKNFGAGM- 150
Query 115 QEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPS 174
L +P +++G+ S+ R +W L + +V VPLYDTLG +
Sbjct 151 --------------LALGARPKQLIGIYSQNRPEWILYEQGCYSFSLVVVPLYDTLGPDA 196
Query 175 VKHIVKLTKMEVLAAEGSKLDAALQLKKEGFPLKAVISF 213
I++ T M+V+ E AA+ L+K LK +++
Sbjct 197 CAFIIRQTDMQVVIVEDDG-KAAMLLEKAPRSLKIIVAI 234
> Hs4503651
Length=699
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 37/179 (20%), Positives = 76/179 (42%), Gaps = 29/179 (16%)
Query 9 KTFKGKYALPIEGTETADSSAVYRAVPDPSNPTQTMLGEPAIPQFPHIKSTYPLLKEAAR 68
K K + L ++ E A S R+ S+ EP + + + + Y + +
Sbjct 49 KPLKPPWHLSMQSVEVAGSGGARRSALLDSD-------EPLVYFYDDVTTLYEGFQRGIQ 101
Query 69 IFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEG 128
+ +G R+P + +++ +Y + E +G AL Q+ G
Sbjct 102 VSNNGPCLGSRKPDQP---------YEWLSYKQVAELSECIGSALIQK-----------G 141
Query 129 LNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVL 187
AP + +G+ ++ R +W +++ +V VPLYDTLG ++ +IV ++ ++
Sbjct 142 FKTAPD--QFIGIFAQNRPEWVIIEQGCFAYSMVIVPLYDTLGNEAITYIVNKAELSLV 198
> Hs11276083
Length=698
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/120 (25%), Positives = 58/120 (48%), Gaps = 19/120 (15%)
Query 74 LYVGERQPIKDSNSQTTLSD------FKYFTYADTLRMVEALGRALDQEVGVPLSHYDEE 127
LY G ++ I+ SN+ L +++ +Y + E +G AL Q+
Sbjct 92 LYEGFQRGIQVSNNGPCLGSRKPDQPYEWLSYKQVAELSECIGSALIQK----------- 140
Query 128 GLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVL 187
G AP + +G+ ++ R +W +++ +V VPLYDTLG ++ +IV ++ ++
Sbjct 141 GFKTAPD--QFIGIFAQNRPEWVIIEQGCFAYSMVIVPLYDTLGNEAITYIVNKAELSLV 198
> CE19275
Length=687
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 32/124 (25%), Positives = 53/124 (42%), Gaps = 13/124 (10%)
Query 92 SDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRL 151
+D+++ TY D + L L E G+ P +G+ +R W +
Sbjct 100 ADYEFLTYDDVHEQAKNLSMTLVHEFGL------------TPANTTNIGIYARNSPQWLV 147
Query 152 LDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEGFP-LKAV 210
A + +V VPLYDTLG + I+ ++ V+ + K +L +E P LK +
Sbjct 148 SAVACVEQSMVVVPLYDTLGAEAATFIISQAEISVVIVDSFKKAESLIKNRENMPTLKNI 207
Query 211 ISFD 214
I D
Sbjct 208 IVID 211
> At1g77590
Length=691
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 39/178 (21%), Positives = 73/178 (41%), Gaps = 24/178 (13%)
Query 47 EPAIPQFPHIKSTYPLLKEAARIFGRNLYVGERQ----PIKDSNSQTT-----LSDFKYF 97
EP + HI + L + + +++G R+ I+ S T L D+++
Sbjct 48 EPVSSHWEHISTLPELFEISCNAHSDRVFLGTRKLISREIETSEDGKTFEKLHLGDYEWL 107
Query 98 TYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAAN 157
T+ TL V L V + H EE RV + + TR +W +
Sbjct 108 TFGKTLEAVCDFASGL-----VQIGHKTEE---------RV-AIFADTREEWFISLQGCF 152
Query 158 YRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEGFPLKAVISFDE 215
R + V +Y +LGE ++ H + T++ + +L + + ++ +K VI D+
Sbjct 153 RRNVTVVTIYSSLGEEALCHSLNETEVTTVICGSKELKKLMDISQQLETVKRVICMDD 210
> At1g64400
Length=665
Score = 42.4 bits (98), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 35/125 (28%), Positives = 60/125 (48%), Gaps = 16/125 (12%)
Query 94 FKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLD 153
+ + TY + +V LG ++ + +GV D+ G+ A P +W +
Sbjct 77 YVWQTYKEVHNVVIKLGNSI-RTIGVGKG--DKCGIYGANSP------------EWIISM 121
Query 154 FAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLK-KEGFPLKAVIS 212
A N G+ VPLYDTLG +++ I+ ++ + AE +K+ L+ K LK ++S
Sbjct 122 EACNAHGLYCVPLYDTLGAGAIEFIICHAEVSLAFAEENKISELLKTAPKSTKYLKYIVS 181
Query 213 FDEPT 217
F E T
Sbjct 182 FGEVT 186
> Hs17505198
Length=697
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 36/162 (22%), Positives = 69/162 (42%), Gaps = 27/162 (16%)
Query 57 KSTYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQE 116
++ Y + + I G +G R+P + +++ +Y + E LG L Q
Sbjct 90 RTMYQVFRRGLSISGNGPCLGFRKPKQP---------YQWLSYQEVADRAEFLGSGLLQH 140
Query 117 VGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVK 176
N + +G+ ++ R +W +++ A +V VPLYDTLG +++
Sbjct 141 -------------NCKACTDQFIGVFAQNRPEWIIVELACYTYSMVVVPLYDTLGPGAIR 187
Query 177 HIVKLTKMEVLAAEGSKLDAALQL----KKEGFPLKAVISFD 214
+I+ + + + + A L L +KE LK +I D
Sbjct 188 YIINTADISTVIVDKPQ-KAVLLLEHVERKETPGLKLIILMD 228
> At2g47240
Length=660
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 48/209 (22%), Positives = 86/209 (41%), Gaps = 33/209 (15%)
Query 8 VKTFKGKYALPIEGTETADSSA-VYRAVPDPSNPTQTMLGEPAIPQF-PHIKSTYPLLKE 65
+K+F K ++G + S VYR +L E P I + + + +
Sbjct 1 MKSFAAKVEEGVKGIDGKPSVGPVYR----------NLLSEKGFPPIDSEITTAWDIFSK 50
Query 66 AARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYD 125
+ F N +G R+ + + + + + TY + V +G AL P S
Sbjct 51 SVEKFPDNNMLGWRRIVDEK-----VGPYMWKTYKEVYEEVLQIGSALRAAGAEPGSRVG 105
Query 126 EEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKME 185
G+N P+ W + A ++ VPLYDTLG +V +IV+ +++
Sbjct 106 IYGVN-CPQ--------------WIIAMEACAAHTLICVPLYDTLGSGAVDYIVEHAEID 150
Query 186 VLAAEGSKLDAALQLK-KEGFPLKAVISF 213
+ + +K+ L+ K LKA++SF
Sbjct 151 FVFVQDTKIKGLLEPDCKCAKRLKAIVSF 179
> ECU10g0890
Length=626
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 38/80 (47%), Gaps = 4/80 (5%)
Query 139 LGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSK----L 194
+G+ S R +W + + A+ G + VPLY T V+ I+ T+M+V A K L
Sbjct 88 VGIYSVNRYEWVVSEMASYMSGCINVPLYSTFSPADVRQILAETEMKVCVASADKAQLLL 147
Query 195 DAALQLKKEGFPLKAVISFD 214
+ L+ K G ++ D
Sbjct 148 ETVLEDSKAGLSCIVLMDRD 167
> CE24550
Length=719
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 37/153 (24%), Positives = 72/153 (47%), Gaps = 26/153 (16%)
Query 71 GRNLYVGERQPIKDSNSQTTL------SD----FKYFTYADTLRMVEALGRALDQEVGVP 120
R LY G R+ + SN+ L SD + + +Y L + + A +E+GVP
Sbjct 108 ARTLYQGVRRGARLSNNGPMLGRRVKQSDGSIPYVWESYNTILERADNVSVAF-RELGVP 166
Query 121 LSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAA-NYRGIVTVPLYDTLGEPSVKHIV 179
+ + +G+ S+ R +W + +FA NY ++ VP+Y+TLG + I+
Sbjct 167 TGNAEN------------IGIYSKNRAEWIITEFATYNYSNVI-VPIYETLGSEASIFIL 213
Query 180 KLTKMEVLAAEG-SKLDAALQLKKEGFPLKAVI 211
+++++ + SK L+ K++ L ++
Sbjct 214 NQAEIKIVVCDDISKATGLLKFKEQCPSLSTLV 246
> ECU10g0910
Length=708
Score = 40.0 bits (92), Expect = 0.003, Method: Composition-based stats.
Identities = 22/71 (30%), Positives = 35/71 (49%), Gaps = 4/71 (5%)
Query 139 LGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSK----L 194
+G+ S R +W + + A+ G + VPLY T V+ I+ T+M+V A K L
Sbjct 170 VGIYSVNRYEWVVSEMASYMSGCINVPLYSTFSPADVRQILAETEMKVCVASADKAQLLL 229
Query 195 DAALQLKKEGF 205
+ L+ K G
Sbjct 230 ETVLEDSKAGL 240
> Hs7706449
Length=683
Score = 40.0 bits (92), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 29/127 (22%), Positives = 52/127 (40%), Gaps = 15/127 (11%)
Query 64 KEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSH 123
K +F R L V + P +++ +Y E LG L +
Sbjct 75 KTMYEVFQRGLAVSDNGPCLGYRKPN--QPYRWLSYKQVSDRAEYLGSCLLHK------- 125
Query 124 YDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTK 183
G +P + +G+ ++ R +W + + A +V VPLYDTLG ++ HIV
Sbjct 126 ----GYKSSPD--QFVGIFAQNRPEWIISELACYTYSMVAVPLYDTLGPEAIVHIVNKAD 179
Query 184 MEVLAAE 190
+ ++ +
Sbjct 180 IAMVICD 186
> CE29312
Length=731
Score = 38.9 bits (89), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 40/189 (21%), Positives = 75/189 (39%), Gaps = 24/189 (12%)
Query 34 VPDPSNPTQTMLGEPAIPQFPHIKSTYPLLKEAARIFGRNLYVGERQPIKDSNSQT---- 89
V D P + + G A FP + + ++ ++ + +G RQ I+ +
Sbjct 71 VDDRDGPIRNIQGH-ASETFPGKDTVDKVWRKCVELYDESPCLGTRQLIEVHEDKQPGGR 129
Query 90 -----TLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSR 144
L ++ + +Y + V L + L+H ++ NP + + +
Sbjct 130 VFEKWHLGEYVWQSYKEVETQVARLAAGIKD-----LAHEEQ---NPK------VVIFAE 175
Query 145 TRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEG 204
TR DW + A + V +Y TLGE ++ H + T+ +L L + L K+
Sbjct 176 TRADWLITALACFRANVTIVTVYATLGEDAIAHAIGETEATILVTSSELLPKIVTLGKKC 235
Query 205 FPLKAVISF 213
LK +I F
Sbjct 236 PTLKTLIYF 244
> At1g49430
Length=666
Score = 38.9 bits (89), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 33/161 (20%), Positives = 66/161 (40%), Gaps = 21/161 (13%)
Query 56 IKSTYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQ 115
I S + EA + + +G+R + + + + + TY + +G A+
Sbjct 44 IDSPWQFFSEAVKKYPNEQMLGQR-----VTTDSKVGPYTWITYKEAHDAAIRIGSAIRS 98
Query 116 EVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSV 175
G++P G+ +W + A +GI VPLYD+LG +V
Sbjct 99 R-----------GVDPG----HCCGIYGANCPEWIIAMEACMSQGITYVPLYDSLGVNAV 143
Query 176 KHIVKLTKMEVLAAEGSKLDAALQLKKEGFP-LKAVISFDE 215
+ I+ ++ ++ + + + L +K LK ++SF E
Sbjct 144 EFIINHAEVSLVFVQEKTVSSILSCQKGCSSNLKTIVSFGE 184
> YER015w
Length=744
Score = 38.9 bits (89), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 35/163 (21%), Positives = 71/163 (43%), Gaps = 9/163 (5%)
Query 55 HIKSTYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALD 114
++++ Y +AR + + +G R PI D + T F++ +Y+ + +G +
Sbjct 91 NLRTAYDHFMFSARRWPQRDCLGSR-PI-DKATGTWEETFRFESYSTVSKRCHNIGSGI- 147
Query 115 QEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPS 174
LS + + P V+ + S +W L D A + LY+TLG +
Sbjct 148 ------LSLVNTKRKRPLEANDFVVAILSHNNPEWILTDLACQAYSLTNTALYETLGPNT 201
Query 175 VKHIVKLTKMEVLAAEGSKLDAALQLKKEGFPLKAVISFDEPT 217
++I+ LT+ +L S + L++ + + ++ DE T
Sbjct 202 SEYILNLTEAPILIFAKSNMYHVLKMVPDMKFVNTLVCMDELT 244
> CE06286
Length=480
Score = 38.5 bits (88), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 31/52 (59%), Gaps = 0/52 (0%)
Query 139 LGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAE 190
+G+ S+ R +W L D A + V+VPLYDT+ + +I L ++ ++ A+
Sbjct 148 IGIYSKNRPEWILSDMAIHNFSNVSVPLYDTITNDDMHYITNLCEISLIFAD 199
> YOR317w
Length=700
Score = 37.7 bits (86), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 31/124 (25%), Positives = 55/124 (44%), Gaps = 13/124 (10%)
Query 91 LSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWR 150
LS + Y ++ ++ +GR L V + L D++ L+ L + T W
Sbjct 94 LSHYHYNSFDQLTDIMHEIGRGL---VKIGLKPNDDDKLH----------LYAATSHKWM 140
Query 151 LLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEGFPLKAV 210
+ A +GI V YDTLGE + H + T + + + S L + ++ + +K +
Sbjct 141 KMFLGAQSQGIPVVTAYDTLGEKGLIHSLVQTGSKAIFTDNSLLPSLIKPVQAAQDVKYI 200
Query 211 ISFD 214
I FD
Sbjct 201 IHFD 204
> CE23909
Length=700
Score = 37.4 bits (85), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 21/80 (26%), Positives = 41/80 (51%), Gaps = 1/80 (1%)
Query 139 LGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGS-KLDAA 197
+G+ S R +W L + A + V+VPLYDT+ + +I L ++ ++ + K
Sbjct 144 IGIYSNNRPEWILSEMAIHNFSNVSVPLYDTITNDDMHYITNLCEISLMFVDAEIKTKQL 203
Query 198 LQLKKEGFPLKAVISFDEPT 217
++ K LK ++ F+E +
Sbjct 204 IRDKSYLSSLKYIVQFNECS 223
> At4g23850
Length=666
Score = 37.4 bits (85), Expect = 0.025, Method: Composition-based stats.
Identities = 50/214 (23%), Positives = 86/214 (40%), Gaps = 51/214 (23%)
Query 14 KYALPIE-GTETADS----SAVYRAV------PDPSNPTQTMLGEPAIPQFPHIKSTYPL 62
KY +E G E +D VYR++ PDP + S + +
Sbjct 6 KYIFQVEEGKEGSDGRPSVGPVYRSIFAKDGFPDP---------------IEGMDSCWDV 50
Query 63 LKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLS 122
+ + + N +G R+ + + + + TY + +V LG +L + VGV
Sbjct 51 FRMSVEKYPNNPMLGRREIVDGKPGK-----YVWQTYQEVYDIVMKLGNSL-RSVGVK-- 102
Query 123 HYDEE--GLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVK 180
DE G+ A P +W + A N G+ VPLYDTLG +V+ I+
Sbjct 103 --DEAKCGIYGANSP------------EWIISMEACNAHGLYCVPLYDTLGADAVEFIIS 148
Query 181 LTKMEVLAAEGSKLDAALQLKKEGFP-LKAVISF 213
+++ ++ E K+ + +K V+SF
Sbjct 149 HSEVSIVFVEEKKISELFKTCPNSTEYMKTVVSF 182
> CE12528
Length=723
Score = 37.4 bits (85), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 22/81 (27%), Positives = 41/81 (50%), Gaps = 6/81 (7%)
Query 139 LGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAAL 198
+G+ S+ R +W L D A + V+VP+YD + + +I L ++ ++ + D
Sbjct 140 IGIYSKNRPEWVLSDMAIHNFSNVSVPIYDNIPNEDMHYITNLCEIPLMFVDSE--DKTT 197
Query 199 QLKKE----GFPLKAVISFDE 215
QL K+ LK ++ FD+
Sbjct 198 QLIKDKTYLSKSLKYIVQFDK 218
> 7304019
Length=766
Score = 36.2 bits (82), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 34/162 (20%), Positives = 61/162 (37%), Gaps = 31/162 (19%)
Query 26 DSSAVYRAVPDPSNPTQTMLGEPAIPQFPHIKSTYPLLKEAARIFGRNLYVGERQPIKDS 85
D+ YR P + ML E +I + + A+ + +G RQ + +
Sbjct 61 DNELTYRTTDPPRDVHVKMLQE-------NIDTLEKVFNYVAKTYTSKRCLGTRQILSEE 113
Query 86 NS---------QTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPL 136
+ + L D+K+ T+ + R GR L +E+G KP
Sbjct 114 DEVQQNGRVFKKYNLGDYKWKTFTEAERTAANFGRGL-RELG--------------QKPR 158
Query 137 RVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHI 178
+ + + TR +W + + + V +Y TLG+ V H
Sbjct 159 ENIVIFAETRAEWMIAAHGCFKQAMPIVTVYATLGDDGVAHC 200
> SPBC18H10.02
Length=676
Score = 35.8 bits (81), Expect = 0.068, Method: Compositional matrix adjust.
Identities = 39/139 (28%), Positives = 57/139 (41%), Gaps = 23/139 (16%)
Query 77 GERQPIKDSNSQTTLSDFKYFTYAD----TLRMVEALGRALDQEVGVPLSHYDEEGLNPA 132
GE++ + S S LSD+ Y ++ D LR AL R L GLN
Sbjct 77 GEKKEVPKSWSYFELSDYNYLSFNDIYDKALRYAGAL-RKL--------------GLNKG 121
Query 133 PKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGS 192
K L + T W L A + + V YDTLGE + H ++ + + + EG
Sbjct 122 DK----FELYAPTSAFWLLTAEACLSQSMTIVTAYDTLGEEGLLHSLRESGVRGMYTEGH 177
Query 193 KLDAALQLKKEGFPLKAVI 211
L + KE L+ +I
Sbjct 178 LLKTLVNPLKEIESLEVII 196
> At4g11030
Length=666
Score = 35.4 bits (80), Expect = 0.077, Method: Composition-based stats.
Identities = 40/166 (24%), Positives = 71/166 (42%), Gaps = 25/166 (15%)
Query 53 FPH----IKSTYPLLKEAARIFGRNLYVGERQPIKDSNSQTTLSDFKYFTYADTLRMVEA 108
FP+ I+S + + + A + N +G R+ SN + + + TY + +V
Sbjct 37 FPNPIDGIQSCWDIFRTAVEKYPNNRMLGRREI---SNGKA--GKYVWKTYKEVYDIVIK 91
Query 109 LGRALDQEVGVPLSHYDEEGLNPAPKPLRVLGLRSRTRMDWRLLDFAANYRGIVTVPLYD 168
LG +L + G+ +EG G+ +W + A N G+ VPLYD
Sbjct 92 LGNSL-RSCGI------KEG--------EKCGIYGINCCEWIISMEACNAHGLYCVPLYD 136
Query 169 TLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEGFP-LKAVISF 213
TLG +V+ I+ ++ + E K+ + +K V+SF
Sbjct 137 TLGAGAVEFIISHAEVSIAFVEEKKIPELFKTCPNSTKYMKTVVSF 182
> 7301111
Length=544
Score = 32.0 bits (71), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 19/76 (25%), Positives = 36/76 (47%), Gaps = 0/76 (0%)
Query 138 VLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAA 197
V+GL S +++ L FA G PL T + V H + L+K +++ A +D
Sbjct 79 VVGLSSENSVNFALAMFAGLAVGATVAPLNVTYSDREVDHAINLSKPKIIFASKITIDRV 138
Query 198 LQLKKEGFPLKAVISF 213
++ + +K +I+
Sbjct 139 AKVASKNKFVKGIIAL 154
> Hs4758330
Length=720
Score = 31.2 bits (69), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 25/107 (23%), Positives = 39/107 (36%), Gaps = 15/107 (14%)
Query 78 ERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLR 137
E QP + L + + +Y D G L Q +G KP
Sbjct 116 EVQPNGKIFKKVILGQYNWLSYEDVFVRAFNFGNGL-QMLG--------------QKPKT 160
Query 138 VLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKM 184
+ + TR +W + A V LY TLG P++ H + T++
Sbjct 161 NIAIFCETRAEWMIAAQACFMYNFQLVTLYATLGGPAIVHALNETEV 207
> 7298132
Length=681
Score = 30.8 bits (68), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 32/70 (45%), Gaps = 1/70 (1%)
Query 148 DWRLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEGFP- 206
+W +F A G V +Y + +V H++ + V + ++ A L+ KE P
Sbjct 106 EWFFAEFGALRAGAVVAGVYPSNSAEAVHHVLATGESSVCVVDDAQQMAKLRAIKERLPR 165
Query 207 LKAVISFDEP 216
LKAVI P
Sbjct 166 LKAVIQLHGP 175
> At1g53090_2
Length=408
Score = 30.8 bits (68), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 29/58 (50%), Gaps = 1/58 (1%)
Query 150 RLLDFAANYRGIVTVPLYDTLGEPSVKHIVKLTKMEVLAAEGSKLDAALQLKKEGFPL 207
+L D + + GI PL+ +G +VK+ V L+ + A GS+ + K FP+
Sbjct 300 KLWDLSMSISGINETPLHSFMGHTNVKNFVGLSVSDGYIATGSETNEVFVYHK-AFPM 356
> 7292211
Length=1180
Score = 30.0 bits (66), Expect = 4.1, Method: Composition-based stats.
Identities = 23/87 (26%), Positives = 39/87 (44%), Gaps = 9/87 (10%)
Query 86 NSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPLRV------- 138
N++ ++++ T LR+ + GRA+ VG+ HYD G NP R+
Sbjct 1064 NTENMQTEYEGSTDYGMLRIGISHGRAMAGVVGISKPHYDIWG-NPVNMASRMDSTGVPG 1122
Query 139 -LGLRSRTRMDWRLLDFAANYRGIVTV 164
+ + T + R + NYRG+ V
Sbjct 1123 QIQVTENTALKLREFNIQCNYRGMTFV 1149
> YIL009w
Length=694
Score = 29.3 bits (64), Expect = 7.2, Method: Composition-based stats.
Identities = 23/101 (22%), Positives = 42/101 (41%), Gaps = 13/101 (12%)
Query 77 GERQPIKDSNSQTTLSDFKYFTYADTLRMVEALGRALDQEVGVPLSHYDEEGLNPAPKPL 136
G+ + I+ + +S +K TY + + ++ +GR L ++G+ P
Sbjct 80 GKDKSIEKTWLYYEMSPYKMMTYQELIWVMHDMGRGL-AKIGI------------KPNGE 126
Query 137 RVLGLRSRTRMDWRLLDFAANYRGIVTVPLYDTLGEPSVKH 177
+ + T W + +GI V YDTLGE + H
Sbjct 127 HKFHIFASTSHKWMKIFLGCISQGIPVVTAYDTLGESGLIH 167
Lambda K H
0.317 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4040519078
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40