bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0156_orf1
Length=144
Score E
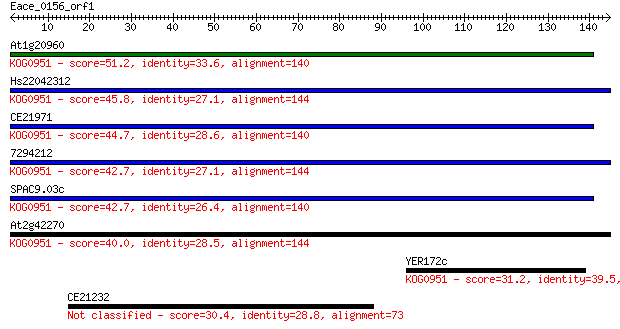
Sequences producing significant alignments: (Bits) Value
At1g20960 51.2 6e-07
Hs22042312 45.8 3e-05
CE21971 44.7 7e-05
7294212 42.7 2e-04
SPAC9.03c 42.7 2e-04
At2g42270 40.0 0.002
YER172c 31.2 0.73
CE21232 30.4 1.1
> At1g20960
Length=2171
Score = 51.2 bits (121), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 47/144 (32%), Positives = 69/144 (47%), Gaps = 24/144 (16%)
Query 1 RWKLFYCIRLGQAQTTEEKDGIREEMKNTQEGQEVLELLDALTSRRNKEKEVTLNVRK-- 58
R K+ +C RL +A+ EE++ I EEM+ G E+ +++ L + R KE N++K
Sbjct 338 RLKVVWCTRLARAEDQEERNRIEEEMRGL--GPELTAIVEQLHATRATAKEREENLQKSI 395
Query 59 --EAATLAAKAQARDASLRAAEADDDSTALGGGAHGTPGGPGGPSSQAAQRKPTGSVDLQ 116
EA L + R AD DS + G G QR+ +DL+
Sbjct 396 NEEARRLKDETGGDGGRGRRDVADRDSES--GWVKG-------------QRQ---MLDLE 437
Query 117 NIAFQQGSHLMANAKVKLPDGTQR 140
++AF QG LMAN K LP G+ R
Sbjct 438 SLAFDQGGLLMANKKCDLPPGSYR 461
> Hs22042312
Length=2136
Score = 45.8 bits (107), Expect = 3e-05, Method: Composition-based stats.
Identities = 39/144 (27%), Positives = 57/144 (39%), Gaps = 28/144 (19%)
Query 1 RWKLFYCIRLGQAQTTEEKDGIREEMKNTQEGQEVLELLDALTSRRNKEKEVTLNVRKEA 60
R + YC L AQ+ EK+ I +M+ E + L L EKE L + +
Sbjct 325 RMMILYCTLLASAQSEAEKERIMGKMEADPELSKFLYQL------HETEKE-DLIREERS 377
Query 61 ATLAAKAQARDASLRAAEADDDSTALGGGAHGTPGGPGGPSSQAAQRKPTGSVDLQNIAF 120
+ D L + D AL P +DL+++ F
Sbjct 378 RRERVRQSRMDTDLETMDLDQGGEALA---------------------PRQVLDLEDLVF 416
Query 121 QQGSHLMANAKVKLPDGTQRIETK 144
QGSH MAN + +LPDG+ R + K
Sbjct 417 TQGSHFMANKRCQLPDGSFRRQRK 440
> CE21971
Length=2145
Score = 44.7 bits (104), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 40/142 (28%), Positives = 63/142 (44%), Gaps = 35/142 (24%)
Query 1 RWKLFYCIRLGQAQTTEEKDGIREEMKNTQEGQEVLELLDALTSRRNKEKEVTLNVRKEA 60
R + YC L QA +E+ I ++M++ E +L LL E
Sbjct 322 RLMILYCTLLRQA-NEKERLQIEDDMRSRPELHPILALLQE---------------TDEG 365
Query 61 ATLAAKAQARDA--SLRAAEADDDSTALGGGAHGTPGGPGGPSSQAAQRKPTGSVDLQNI 118
+ + + RDA S +AA A +++ + G G RK +DL ++
Sbjct 366 SVVQVEKSKRDAEKSKKAATAANEAISAGQWQAG--------------RK---MLDLNDL 408
Query 119 AFQQGSHLMANAKVKLPDGTQR 140
F QGSHLM+N + +LPDG+ R
Sbjct 409 TFSQGSHLMSNKRCELPDGSYR 430
> 7294212
Length=678
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 39/149 (26%), Positives = 56/149 (37%), Gaps = 43/149 (28%)
Query 1 RWKLFYCIRLGQAQTTEEKDGIREEMKNTQEGQEVLELLDALTSRRNKEKEVTLNVRKEA 60
R + YC L AQT E+ IRE+M+ ++L LD S +E
Sbjct 330 RQMVLYCTMLASAQTDSERQRIREKMRGNSALAKILRQLDTGKSEDQEE----------- 378
Query 61 ATLAAKAQARDASLRAAEADDDSTALGGGAHGTPGGPGGPSSQAAQRKPTGS-----VDL 115
G A G+ G G A + ++L
Sbjct 379 ---------------------------GEARGSKRGKGDAEDGGAAAAGQVAGVRQLLEL 411
Query 116 QNIAFQQGSHLMANAKVKLPDGTQRIETK 144
+ +AF QGSH MAN + +LPDG+ R + K
Sbjct 412 EEMAFTQGSHFMANKRCQLPDGSYRKQRK 440
> SPAC9.03c
Length=2176
Score = 42.7 bits (99), Expect = 2e-04, Method: Composition-based stats.
Identities = 37/142 (26%), Positives = 58/142 (40%), Gaps = 31/142 (21%)
Query 1 RWKLFYCIRLGQAQTTEEKDGIREEMKNTQEGQEVLELL--DALTSRRNKEKEVTLNVRK 58
RW + C L +A T EE+ G+ E+++ +LE L A+T + E+ NV +
Sbjct 363 RWTIVSCTMLKRAATDEERLGVEEQIRAAGRSW-ILEALRPGAITIPDDGLNELNNNVVE 421
Query 59 EAATLAAKAQARDASLRAAEADDDSTALGGGAHGTPGGPGGPSSQAAQRKPTGSVDLQNI 118
+A +L + + P VDL+N
Sbjct 422 KAEPAPVSEIPLSKTLTSHKI----------------------------VPKHQVDLENY 453
Query 119 AFQQGSHLMANAKVKLPDGTQR 140
F +GS LM+N VKLP+G+ R
Sbjct 454 VFTEGSRLMSNKAVKLPEGSFR 475
> At2g42270
Length=2172
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 41/148 (27%), Positives = 66/148 (44%), Gaps = 23/148 (15%)
Query 1 RWKLFYCIRLGQAQTTEEKDGIREEMKNTQEGQEVLELLDALTSRR----NKEKEVTLNV 56
R K+ +C RL + + EE++ I EEM G E+ ++ L ++R +E++ ++
Sbjct 338 RLKVVWCTRLARGRDQEERNQIEEEMLGL--GSELAAIVKELHAKRATAKEREEKREKDI 395
Query 57 RKEAATLAAKAQARDASLRAAEADDDSTALGGGAHGTPGGPGGPSSQAAQRKPTGSVDLQ 116
++EA L D R DD G G QR+ +DL+
Sbjct 396 KEEAQHLMDDDSGVDGD-RGMRDVDDLDLENGWLKG-------------QRQ---VMDLE 438
Query 117 NIAFQQGSHLMANAKVKLPDGTQRIETK 144
++AF QG N K +LPD + RI K
Sbjct 439 SLAFNQGGFTRENNKCELPDRSFRIRGK 466
> YER172c
Length=2163
Score = 31.2 bits (69), Expect = 0.73, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 24/49 (48%), Gaps = 6/49 (12%)
Query 96 GPGGPSSQAAQRKPTGS------VDLQNIAFQQGSHLMANAKVKLPDGT 138
G P S A+R + +DL+ I F + S LM KV LP+G+
Sbjct 404 GDDQPQSSEAKRTKFSNPAIPPVIDLEKIKFDESSKLMTVTKVSLPEGS 452
> CE21232
Length=184
Score = 30.4 bits (67), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 35/73 (47%), Gaps = 13/73 (17%)
Query 15 TTEEKDGIREEMKNTQEGQEVLELLDALTSRRNKEKEVTLNVRKEAATLAAKAQARDASL 74
TTEEK I+E +K+ G + +E E++ ++++ + +L AK + D L
Sbjct 50 TTEEKAAIKEFIKSVMGGNKSVE-------------ELSADIKERSPSLYAKVEKLDVLL 96
Query 75 RAAEADDDSTALG 87
R A D AL
Sbjct 97 RTKLAKLDPAALA 109
Lambda K H
0.309 0.126 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1675978996
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40