bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0153_orf1
Length=207
Score E
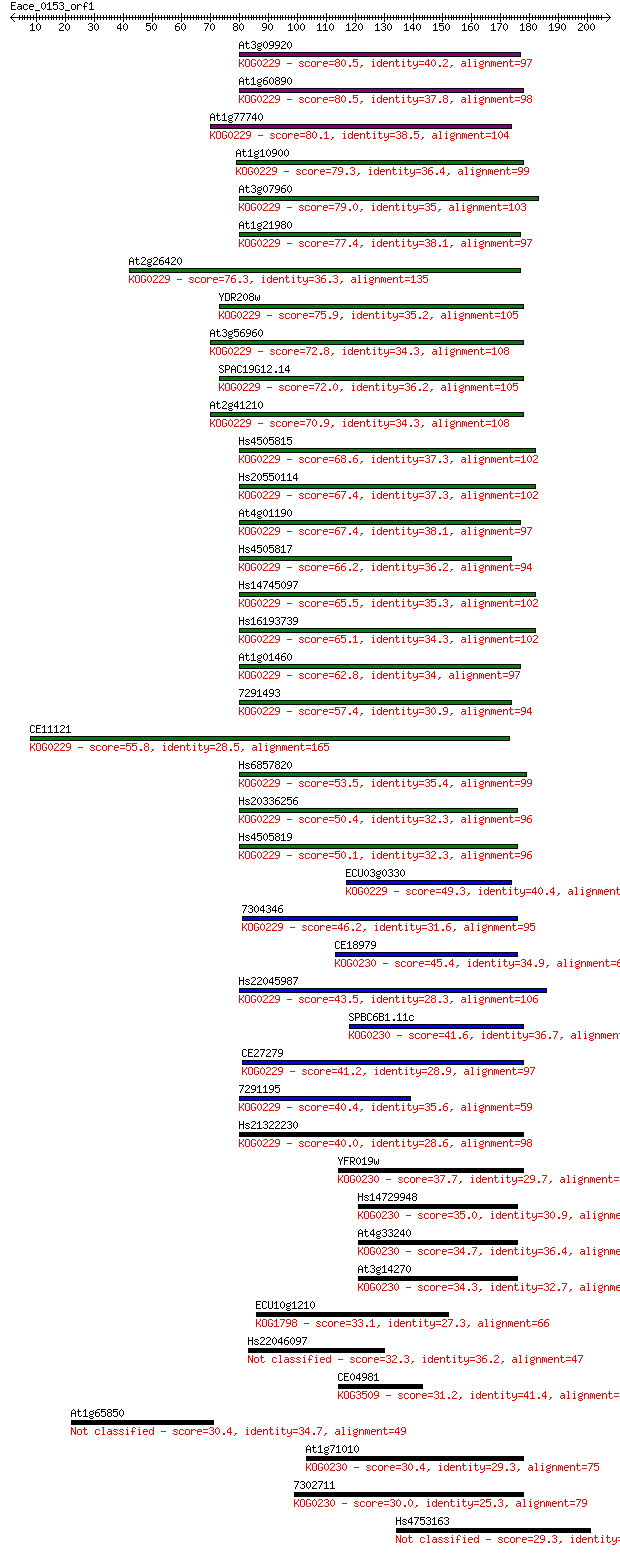
Sequences producing significant alignments: (Bits) Value
At3g09920 80.5 2e-15
At1g60890 80.5 2e-15
At1g77740 80.1 3e-15
At1g10900 79.3 5e-15
At3g07960 79.0 7e-15
At1g21980 77.4 2e-14
At2g26420 76.3 4e-14
YDR208w 75.9 5e-14
At3g56960 72.8 5e-13
SPAC19G12.14 72.0 8e-13
At2g41210 70.9 2e-12
Hs4505815 68.6 8e-12
Hs20550114 67.4 2e-11
At4g01190 67.4 2e-11
Hs4505817 66.2 4e-11
Hs14745097 65.5 8e-11
Hs16193739 65.1 9e-11
At1g01460 62.8 5e-10
7291493 57.4 2e-08
CE11121 55.8 6e-08
Hs6857820 53.5 3e-07
Hs20336256 50.4 3e-06
Hs4505819 50.1 3e-06
ECU03g0330 49.3 5e-06
7304346 46.2 4e-05
CE18979 45.4 9e-05
Hs22045987 43.5 3e-04
SPBC6B1.11c 41.6 0.001
CE27279 41.2 0.002
7291195 40.4 0.003
Hs21322230 40.0 0.003
YFR019w 37.7 0.017
Hs14729948 35.0 0.092
At4g33240 34.7 0.13
At3g14270 34.3 0.19
ECU10g1210 33.1 0.44
Hs22046097 32.3 0.62
CE04981 31.2 1.6
At1g65850 30.4 2.4
At1g71010 30.4 2.7
7302711 30.0 3.2
Hs4753163 29.3 5.4
> At3g09920
Length=815
Score = 80.5 bits (197), Expect = 2e-15, Method: Composition-based stats.
Identities = 39/97 (40%), Positives = 57/97 (58%), Gaps = 1/97 (1%)
Query 80 FQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKTI 139
++D P F RE+ I DY S+C D E S KSG FF S D +F+IKT+
Sbjct 453 WKDYCPMVFRNLREMFKIDAADYMMSICGND-TLRELSSPGKSGSVFFLSQDDRFMIKTL 511
Query 140 SKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIHRVE 176
K+EV +LR+LP Y H+ + +L+T+ FG+HR++
Sbjct 512 RKSEVKVLLRMLPDYHHHVKTYENTLITKFFGLHRIK 548
> At1g60890
Length=769
Score = 80.5 bits (197), Expect = 2e-15, Method: Composition-based stats.
Identities = 37/98 (37%), Positives = 59/98 (60%), Gaps = 1/98 (1%)
Query 80 FQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKTI 139
++D P F RE+ + DY S+C D E S KSG F+ SHD +F+IKT+
Sbjct 408 WKDYCPMVFRNLREMFKLDAADYMMSICGDD-GLREISSPGKSGSIFYLSHDDRFVIKTL 466
Query 140 SKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIHRVEL 177
++E+ +LR+LP Y +H+ +L+T+ FG+HR++L
Sbjct 467 KRSELKVLLRMLPRYYEHVGDYENTLITKFFGVHRIKL 504
> At1g77740
Length=754
Score = 80.1 bits (196), Expect = 3e-15, Method: Composition-based stats.
Identities = 40/104 (38%), Positives = 59/104 (56%), Gaps = 3/104 (2%)
Query 70 PLLSSSFVATFQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFS 129
P LS F ++D P F RE+ + DY ++C D E S KSG FF+ +
Sbjct 406 PHLSVDF--RWKDYCPLVFRRLRELFTVDPADYMLAICGND-ALRELSSPGKSGSFFYLT 462
Query 130 HDGKFLIKTISKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIH 173
D +F+IKT+ K+EV +LR+LPSY H+ +L+TR +G+H
Sbjct 463 QDDRFMIKTVKKSEVKVLLRMLPSYYKHVCQYENTLVTRFYGVH 506
> At1g10900
Length=859
Score = 79.3 bits (194), Expect = 5e-15, Method: Composition-based stats.
Identities = 36/99 (36%), Positives = 59/99 (59%), Gaps = 1/99 (1%)
Query 79 TFQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKT 138
+++D P F R++ + +Y S+C D E S KSG F+ SHD +F+IKT
Sbjct 495 SWKDYCPMVFRNLRQMFKLDAAEYMMSICGDD-GLTEISSPGKSGSIFYLSHDDRFVIKT 553
Query 139 ISKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIHRVEL 177
+ K+E+ +LR+LP Y +H+ +L+T+ FG+HR+ L
Sbjct 554 LKKSELQVLLRMLPKYYEHVGDHENTLITKFFGVHRITL 592
> At3g07960
Length=715
Score = 79.0 bits (193), Expect = 7e-15, Method: Composition-based stats.
Identities = 36/103 (34%), Positives = 62/103 (60%), Gaps = 1/103 (0%)
Query 80 FQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKTI 139
++D P F R++ + DY S+C D E S KSG FF+ ++D +++IKT+
Sbjct 387 WKDYCPVVFRTLRKLFSVDAADYMLSICGND-ALRELSSPGKSGSFFYLTNDDRYMIKTM 445
Query 140 SKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIHRVELYTTNE 182
KAE ++R+LP+Y +H+ + +L+T+ FG+H V+L T +
Sbjct 446 KKAETKVLIRMLPAYYNHVRACENTLVTKFFGLHCVKLTGTAQ 488
> At1g21980
Length=752
Score = 77.4 bits (189), Expect = 2e-14, Method: Composition-based stats.
Identities = 37/97 (38%), Positives = 56/97 (57%), Gaps = 1/97 (1%)
Query 80 FQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKTI 139
++D P F RE+ + Y ++C D E S KSG FF+ + D +F+IKT+
Sbjct 412 WKDYCPLVFRRLRELFQVDPAKYMLAICGND-ALRELSSPGKSGSFFYLTQDDRFMIKTV 470
Query 140 SKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIHRVE 176
K+EV +LR+LPSY H+ SL+TR +G+H V+
Sbjct 471 KKSEVKVLLRMLPSYYKHVCQYENSLVTRFYGVHCVK 507
> At2g26420
Length=705
Score = 76.3 bits (186), Expect = 4e-14, Method: Composition-based stats.
Identities = 49/155 (31%), Positives = 77/155 (49%), Gaps = 23/155 (14%)
Query 42 DGLLLLQLGFLLD----EPLLLEVRHFPF----------------VCSPLLSSSFVATFQ 81
D +L LQLG LL E+RH F P LS+ F ++
Sbjct 330 DLMLNLQLGIRYSVGKHASLLRELRHSDFDPKDKQWTRFPPEGSKSTPPHLSAEF--KWK 387
Query 82 DIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKTISK 141
D P F R++ I DY ++C + EF S KSG F+ + D +++IKT+ K
Sbjct 388 DYCPIVFRHLRDLFAIDQADYMLAICGNE-SLREFASPGKSGSAFYLTQDERYMIKTMKK 446
Query 142 AEVIQILRVLPSYIDHILSTPLSLLTRIFGIHRVE 176
+E+ +L++LP+Y +H+ SL+T+ FG+H V+
Sbjct 447 SEIKVLLKMLPNYYEHVSKYKNSLVTKFFGVHCVK 481
> YDR208w
Length=779
Score = 75.9 bits (185), Expect = 5e-14, Method: Composition-based stats.
Identities = 37/105 (35%), Positives = 60/105 (57%), Gaps = 1/105 (0%)
Query 73 SSSFVATFQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDG 132
SS + F+D P+ F R + G+ DY SL S + E S KSG FF++S D
Sbjct 433 SSQYAFKFKDYCPEVFRELRALFGLDPADYLVSLTSK-YILSELNSPGKSGSFFYYSRDY 491
Query 133 KFLIKTISKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIHRVEL 177
K++IKTI +E I + + + Y +H+ P +L+ + +G+HRV++
Sbjct 492 KYIIKTIHHSEHIHLRKHIQEYYNHVRDNPNTLICQFYGLHRVKM 536
> At3g56960
Length=779
Score = 72.8 bits (177), Expect = 5e-13, Method: Composition-based stats.
Identities = 37/108 (34%), Positives = 62/108 (57%), Gaps = 3/108 (2%)
Query 70 PLLSSSFVATFQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFS 129
P SS F ++D P F R++ + DY S+C D E S KSG FF+ +
Sbjct 440 PHQSSEF--KWKDYCPLVFRSLRKLFKVDPADYMLSICGND-ALRELSSPGKSGSFFYLT 496
Query 130 HDGKFLIKTISKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIHRVEL 177
+D +++IKT+ K+E +LR+L +Y +H+ + +L+ R +G+H V+L
Sbjct 497 NDDRYMIKTMKKSETKVLLRMLAAYYNHVRAFENTLVIRFYGLHCVKL 544
> SPAC19G12.14
Length=742
Score = 72.0 bits (175), Expect = 8e-13, Method: Composition-based stats.
Identities = 38/105 (36%), Positives = 61/105 (58%), Gaps = 1/105 (0%)
Query 73 SSSFVATFQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDG 132
S+ + F+D AP F R++ + DY SL S + E S KSG FF+FS D
Sbjct 316 SAKYDFKFKDYAPWVFRHLRQLFHLDAADYLVSLTSK-YILSELDSPGKSGSFFYFSRDY 374
Query 133 KFLIKTISKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIHRVEL 177
+F+IKTI +E + +L Y +H+ + P +L+++ +G+HRV+L
Sbjct 375 RFIIKTIHHSEHKFLREILYDYYEHVKNNPNTLISQFYGLHRVKL 419
> At2g41210
Length=772
Score = 70.9 bits (172), Expect = 2e-12, Method: Composition-based stats.
Identities = 37/108 (34%), Positives = 61/108 (56%), Gaps = 3/108 (2%)
Query 70 PLLSSSFVATFQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFS 129
P S+ F ++D P F R++ + DY S+C D E S KSG FF+ +
Sbjct 433 PHQSTEF--KWKDYCPLVFRSLRKLFKVDPADYMLSICGND-ALRELSSPGKSGSFFYLT 489
Query 130 HDGKFLIKTISKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIHRVEL 177
+D +++IKT+ K+E +L +L +Y +H+ + SL+ R FG+H V+L
Sbjct 490 NDDRYMIKTMKKSETKVLLGMLAAYYNHVRAFENSLVIRFFGLHCVKL 537
> Hs4505815
Length=549
Score = 68.6 bits (166), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 38/102 (37%), Positives = 56/102 (54%), Gaps = 2/102 (1%)
Query 80 FQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKTI 139
F+ AP AF RE+ GI DY SLCS P IE S+ SG F+ S D +F+IKT+
Sbjct 126 FKTYAPVAFRYFRELFGIRPDDYLYSLCSE--PLIELCSSGASGSLFYVSSDDEFIIKTV 183
Query 140 SKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIHRVELYTTN 181
E + ++LP Y ++ P +LL + +G++ V+ N
Sbjct 184 QHKEAEFLQKLLPGYYMNLNQNPRTLLPKFYGLYCVQAGGKN 225
> Hs20550114
Length=512
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 38/102 (37%), Positives = 55/102 (53%), Gaps = 2/102 (1%)
Query 80 FQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKTI 139
F+ AP AF RE+ GI DY SLCS P IE S+ SG F+ S D + +IKT+
Sbjct 126 FKTYAPVAFRYFRELFGIPPDDYLCSLCSE--PLIELCSSGASGSLFYVSSDDELIIKTL 183
Query 140 SKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIHRVELYTTN 181
E + ++LP Y ++ P +LL + FG++ V+ N
Sbjct 184 QHKEAEFLQKLLPGYYLNLSQNPRTLLPKFFGLYCVQTGGKN 225
> At4g01190
Length=401
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 37/97 (38%), Positives = 54/97 (55%), Gaps = 2/97 (2%)
Query 80 FQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKTI 139
++D P F +E+ GI DY S+C+ + ++ S+ K G F S+D +FLIK +
Sbjct 31 WKDYCPVGFGLIQELEGIDHDDYLLSICTDET--LKKISSGKIGNVFHISNDNRFLIKIL 88
Query 140 SKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIHRVE 176
K+E+ L +LP Y HI SL TRIFG H V+
Sbjct 89 RKSEIKVTLEMLPRYYRHINYHRSSLFTRIFGAHSVK 125
> Hs4505817
Length=540
Score = 66.2 bits (160), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 34/94 (36%), Positives = 52/94 (55%), Gaps = 2/94 (2%)
Query 80 FQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKTI 139
F+ AP AF RE+ GI DY S+CS P IE + SG FF + D +F+IKT+
Sbjct 83 FKTYAPLAFRYFRELFGIKPDDYLYSICSE--PLIELSNPGASGSLFFVTSDDEFIIKTV 140
Query 140 SKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIH 173
E + ++LP Y ++ P +LL + +G++
Sbjct 141 QHKEAEFLQKLLPGYYMNLNQNPRTLLPKFYGLY 174
> Hs14745097
Length=435
Score = 65.5 bits (158), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 36/102 (35%), Positives = 55/102 (53%), Gaps = 2/102 (1%)
Query 80 FQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKTI 139
F+ AP AF E+ GI DY SLCS P IE S+ SG F+ S D +F++KT+
Sbjct 86 FKTYAPVAFRYFWELFGIRPDDYLYSLCSE--PLIELCSSGASGSLFYVSSDDEFIVKTV 143
Query 140 SKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIHRVELYTTN 181
E + ++LP Y ++ P +LL + +G++ V+ N
Sbjct 144 RHKEAEFLQKLLPGYYINLNQNPRTLLPKFYGLYCVQTGGKN 185
> Hs16193739
Length=668
Score = 65.1 bits (157), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 35/102 (34%), Positives = 55/102 (53%), Gaps = 2/102 (1%)
Query 80 FQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKTI 139
F+ AP AF RE+ GI DY SLC+ P IE + SG F+ + D +F+IKT+
Sbjct 133 FKTYAPVAFRYFRELFGIRPDDYLYSLCNE--PLIELSNPGASGSLFYVTSDDEFIIKTV 190
Query 140 SKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIHRVELYTTN 181
E + ++LP Y ++ P +LL + +G++ V+ N
Sbjct 191 MHKEAEFLQKLLPGYYMNLNQNPRTLLPKFYGLYCVQSGGKN 232
> At1g01460
Length=425
Score = 62.8 bits (151), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 33/97 (34%), Positives = 53/97 (54%), Gaps = 2/97 (2%)
Query 80 FQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKTI 139
++D P F +E+ I+ +Y S+C+ + + S SK G F S D +FLIK +
Sbjct 39 WKDYCPLGFRLIQELEDINHDEYMKSICNDET--LRKLSTSKVGNMFLLSKDDRFLIKIL 96
Query 140 SKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIHRVE 176
K+E+ IL +LP Y HI +LL++ +G H V+
Sbjct 97 RKSEIKVILEMLPGYFRHIHKYRSTLLSKNYGAHSVK 133
> 7291493
Length=515
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 52/94 (55%), Gaps = 2/94 (2%)
Query 80 FQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKTI 139
++ AP AF R++ GI D+ S+C++ P E + SG F+ + D +F+IKT+
Sbjct 134 YKIYAPIAFRYFRDLFGIQPDDFMMSMCTS--PLRELSNPGASGSIFYLTTDDEFIIKTV 191
Query 140 SKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIH 173
E + ++LP Y ++ P +LL + FG++
Sbjct 192 QHKEGEFLQKLLPGYYMNLNQNPRTLLPKFFGLY 225
> CE11121
Length=619
Score = 55.8 bits (133), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 47/167 (28%), Positives = 78/167 (46%), Gaps = 20/167 (11%)
Query 8 PSYALTVAMKKGMEFSNMMSAGLVSLAHREGSQLDGLLLLQLGFLLDEPLLLEVRHFPFV 67
P+ AL A++ G+ SN + + L SL +R+ LL + +++ FP
Sbjct 84 PTNALMQAIQLGI--SNSIGS-LASLPNRD-------------VLLQDFEKVDIVAFPAA 127
Query 68 CSPLLSSSFVA--TFQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQF 125
S + S F+ AP AF R + I D+ S+C+ P E + SG
Sbjct 128 GSTITPSHSFGDFRFRTYAPIAFRYFRNLFHIKPADFLRSICTE--PLKELSNAGASGSI 185
Query 126 FFFSHDGKFLIKTISKAEVIQILRVLPSYIDHILSTPLSLLTRIFGI 172
F+ S D +F+IKT+ E + ++LP Y ++ P +LL + FG+
Sbjct 186 FYVSQDDQFIIKTVQHKEADFLQKLLPGYYMNLNQNPRTLLPKFFGL 232
> Hs6857820
Length=406
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 35/106 (33%), Positives = 58/106 (54%), Gaps = 10/106 (9%)
Query 80 FQDIAPDAFECAREVAGISDQDYRTSLC-STDFPFIEFQSNSKSGQFFFFSHDGKFLIKT 138
F++ P F RE GI DQD++ SL S P S ++SG F S+D +++IKT
Sbjct 90 FKEYCPMVFRNLRERFGIDDQDFQNSLTRSAPLPN---DSQARSGARFHTSYDKRYIIKT 146
Query 139 ISKAEVIQILRVLPSYIDHILST-PLSLLTRIFGIHR-----VELY 178
I+ +V ++ +L Y +I+ ++LL + G++R VE+Y
Sbjct 147 ITSEDVAEMHNILKKYHQYIVECHGITLLPQFLGMYRLNVDGVEIY 192
> Hs20336256
Length=281
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 31/97 (31%), Positives = 51/97 (52%), Gaps = 3/97 (3%)
Query 80 FQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKTI 139
F++ P F RE GI DQDY+ S+ T I S + G F ++D +F+IKT+
Sbjct 95 FKEYCPMVFRNLRERFGIDDQDYQNSV--TRSAPINSDSQGRCGTRFLTTYDRRFVIKTV 152
Query 140 SKAEVIQILRVLPSYIDHILST-PLSLLTRIFGIHRV 175
S +V ++ +L Y I+ +LL + G++R+
Sbjct 153 SSEDVAEMHNILKKYHQFIVECHGNTLLPQFLGMYRL 189
> Hs4505819
Length=416
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 31/97 (31%), Positives = 51/97 (52%), Gaps = 3/97 (3%)
Query 80 FQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKTI 139
F++ P F RE GI DQDY+ S+ T I S + G F ++D +F+IKT+
Sbjct 95 FKEYCPMVFRNLRERFGIDDQDYQNSV--TRSAPINSDSQGRCGTRFLTTYDRRFVIKTV 152
Query 140 SKAEVIQILRVLPSYIDHILST-PLSLLTRIFGIHRV 175
S +V ++ +L Y I+ +LL + G++R+
Sbjct 153 SSEDVAEMHNILKKYHQFIVECHGNTLLPQFLGMYRL 189
> ECU03g0330
Length=296
Score = 49.3 bits (116), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 23/57 (40%), Positives = 34/57 (59%), Gaps = 0/57 (0%)
Query 117 QSNSKSGQFFFFSHDGKFLIKTISKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIH 173
Q + KSG F FF+ D KF++KTI K E I ++ Y ++L P+S L +I G +
Sbjct 72 QIHGKSGSFLFFTRDFKFIVKTIRKNEFRCIRGMINEYKVYVLENPMSFLCKILGCY 128
> 7304346
Length=313
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 53/96 (55%), Gaps = 3/96 (3%)
Query 81 QDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKTIS 140
++ P F RE G+ D DYR SL + P I+ S+ KSG F+ S+D F+IK+++
Sbjct 89 KEYCPLVFRNLRERFGVDDVDYRESLTRSQ-P-IQIDSSGKSGAQFYQSYDKFFIIKSLT 146
Query 141 KAEVIQILRVLPSYIDHILSTP-LSLLTRIFGIHRV 175
E+ ++ L Y +++ +LL + G++R+
Sbjct 147 SEEIERMHAFLKQYHPYVVERHGKTLLPQYLGMYRI 182
> CE18979
Length=1375
Score = 45.4 bits (106), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 43/75 (57%), Gaps = 12/75 (16%)
Query 113 FIEFQSNS--------KSGQFFFFSHDGKFLIKTISKAEVIQILRVLPSYIDHILS---- 160
FI SNS KSG FF+ + D +F++K +S+ E+ ++ P+Y D++ +
Sbjct 1126 FIRSLSNSTFWTPQGGKSGSFFYRTQDDRFVVKQMSRFEIQSFVKFAPNYFDYLTTSATE 1185
Query 161 TPLSLLTRIFGIHRV 175
+ L+ L +++G+ R+
Sbjct 1186 SKLTTLCKVYGVFRI 1200
> Hs22045987
Length=255
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 30/106 (28%), Positives = 48/106 (45%), Gaps = 2/106 (1%)
Query 80 FQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKTI 139
F+ P AF +E+ GI D SLCS P IE S+ G S +F+IKT+
Sbjct 101 FKTYTPVAFCYFQELFGICPDDDLYSLCSE--PLIELSSSGADGSLLHVSIHNEFIIKTV 158
Query 140 SKAEVIQILRVLPSYIDHILSTPLSLLTRIFGIHRVELYTTNEAAA 185
+ + +++P Y + +LL + +G+ V+ N A
Sbjct 159 QHKQAEFLQKLIPGYHIDLNQNSWTLLPKFYGLCCVKAGGKNTQIA 204
> SPBC6B1.11c
Length=614
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 36/64 (56%), Gaps = 4/64 (6%)
Query 118 SNSKSGQFFFFSHDGKFLIKTISKAEVIQILRVLPSYIDHILSTPL----SLLTRIFGIH 173
S KSG F + D K+++K +S+ E +L P+Y D+I + LT+IFG +
Sbjct 376 SGGKSGSAFLKTFDKKYILKVLSRLESDSLLNFAPAYFDYISKVFFHELPTALTKIFGFY 435
Query 174 RVEL 177
RV++
Sbjct 436 RVDI 439
> CE27279
Length=401
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 52/99 (52%), Gaps = 2/99 (2%)
Query 81 QDIAPDAFECAREVAGISDQDYRTSLCSTD-FPFIEFQSNSKSGQFFFFSHDGKFLIKTI 139
++ P+ F RE G+ + +Y SL S + P + S S FF S+D KF+IK++
Sbjct 85 KEYCPNVFRNLREQFGVDNFEYLRSLTSYEPEPDLLDGSAKDSTPRFFISYDKKFVIKSM 144
Query 140 SKAEVIQILRVLPSYIDHILSTP-LSLLTRIFGIHRVEL 177
V ++ VL +Y +++ +LL + G++R+ +
Sbjct 145 DSEAVAELHSVLRNYHQYVVEKQGKTLLPQYLGLYRLTI 183
> 7291195
Length=770
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 34/59 (57%), Gaps = 2/59 (3%)
Query 80 FQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKT 138
F+ AP AF R++ GI+ D+ S+C++ P E + SG F+ + D +F+IKT
Sbjct 213 FKVYAPIAFRYFRDLFGIAPDDFLMSMCAS--PLRELSNPGASGSIFYLTTDDEFIIKT 269
> Hs21322230
Length=421
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 47/99 (47%), Gaps = 6/99 (6%)
Query 80 FQDIAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFSHDGKFLIKTI 139
F++ P F R+ GI DQDY SL +S G+ F S+D +IK +
Sbjct 100 FKEYCPQVFRNLRDRFGIDDQDYLVSLTRNP----PSESEGSDGR-FLISYDRTLVIKEV 154
Query 140 SKAEVIQILRVLPSYIDHILST-PLSLLTRIFGIHRVEL 177
S ++ + L +Y +I+ +LL + G++RV +
Sbjct 155 SSEDIADMHSNLSNYHQYIVKCHGNTLLPQFLGMYRVSV 193
> YFR019w
Length=2278
Score = 37.7 bits (86), Expect = 0.017, Method: Composition-based stats.
Identities = 19/69 (27%), Positives = 40/69 (57%), Gaps = 5/69 (7%)
Query 114 IEFQSNS-KSGQFFFFSHDGKFLIKTISKAEVIQILRVLPSYIDHILSTPL----SLLTR 168
+++ SN KSG F + D +F+IK +S AE+ ++ PSY +++ + L +
Sbjct 2035 VKWDSNGGKSGSGFLKTLDDRFIIKELSHAELEAFIKFAPSYFEYMAQAMFHDLPTTLAK 2094
Query 169 IFGIHRVEL 177
+FG +++++
Sbjct 2095 VFGFYQIQV 2103
> Hs14729948
Length=558
Score = 35.0 bits (79), Expect = 0.092, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 32/59 (54%), Gaps = 4/59 (6%)
Query 121 KSGQFFFFSHDGKFLIKTISKAEVIQILRVLPSYIDHILS----TPLSLLTRIFGIHRV 175
KSG F+ + D +F++K + + EV L P Y ++I + + L +I G++R+
Sbjct 321 KSGAAFYATEDDRFILKQMPRLEVQSFLDFAPHYFNYITNAVQQKRPTALAKILGVYRI 379
> At4g33240
Length=1757
Score = 34.7 bits (78), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 34/60 (56%), Gaps = 6/60 (10%)
Query 121 KSGQFFFFSHDGKFLIKTISKAEVIQILRVLPSYIDHI-----LSTPLSLLTRIFGIHRV 175
KS FF S D +F+IK ++K E+ ++ P+Y ++ +P S L +I GI++V
Sbjct 1498 KSNVFFAKSLDDRFIIKQVTKTELESFIKFGPAYFKYLTESISTKSPTS-LAKILGIYQV 1556
> At3g14270
Length=1791
Score = 34.3 bits (77), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 33/59 (55%), Gaps = 4/59 (6%)
Query 121 KSGQFFFFSHDGKFLIKTISKAEVIQILRVLPSYIDH----ILSTPLSLLTRIFGIHRV 175
KS FF + D +F+IK ++K E+ ++ P+Y + I + + L +I GI++V
Sbjct 1537 KSNVFFAKTLDDRFIIKQVTKTELESFIKFAPAYFKYLSESISTKSPTCLAKILGIYQV 1595
> ECU10g1210
Length=1799
Score = 33.1 bits (74), Expect = 0.44, Method: Composition-based stats.
Identities = 18/69 (26%), Positives = 35/69 (50%), Gaps = 3/69 (4%)
Query 86 DAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNS---KSGQFFFFSHDGKFLIKTISKA 142
D ++C E G++ +D+ S+C D P +N + G+++ F + G+ + S+
Sbjct 143 DEYDCFGEGRGLNAEDFIVSICEHDIPVEVCAANDLGIRCGRWYGFRYTGEMYVIEKSER 202
Query 143 EVIQILRVL 151
V LR+L
Sbjct 203 IVFPDLRIL 211
> Hs22046097
Length=145
Score = 32.3 bits (72), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 27/47 (57%), Gaps = 1/47 (2%)
Query 83 IAPDAFECAREVAGISDQDYRTSLCSTDFPFIEFQSNSKSGQFFFFS 129
+A AF R G++++DY+ +L P+++F S SKS FF S
Sbjct 100 LAGPAFAWLRRSLGLAEEDYQAAL-GPGGPYLQFLSTSKSKASFFLS 145
> CE04981
Length=757
Score = 31.2 bits (69), Expect = 1.6, Method: Composition-based stats.
Identities = 12/29 (41%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 114 IEFQSNSKSGQFFFFSHDGKFLIKTISKA 142
I+F++ SG FF+ D KFLI T+ +
Sbjct 419 IQFKTEKPSGVLFFWKEDSKFLIVTVENS 447
> At1g65850
Length=926
Score = 30.4 bits (67), Expect = 2.4, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 24/50 (48%), Gaps = 1/50 (2%)
Query 22 FSNMMSAGLVSLAHREGSQLDGLLLLQ-LGFLLDEPLLLEVRHFPFVCSP 70
F M + + +R G + D L L Q L +L + +LE HFP C P
Sbjct 468 FEGMPNLKFLRFYYRYGDESDKLYLPQGLNYLSQKLKILEWDHFPLTCMP 517
> At1g71010
Length=1609
Score = 30.4 bits (67), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 22/87 (25%), Positives = 38/87 (43%), Gaps = 12/87 (13%)
Query 103 RTSLCSTDFPFIEFQSN--------SKSGQFFFFSHDGKFLIKTISKAEVIQILRVLPSY 154
R + C ++ F+ S KS +F S D +F+IK + K E+ P Y
Sbjct 1353 RKTCCPSEVDFVRSLSRCQRWSAQGGKSNVYFAKSLDERFIIKQVVKTELDSFEDFAPEY 1412
Query 155 IDHIL----STPLSLLTRIFGIHRVEL 177
++ S + L +I GI++V +
Sbjct 1413 FKYLKESLSSGSPTCLAKILGIYQVSI 1439
> 7302711
Length=1809
Score = 30.0 bits (66), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 20/84 (23%), Positives = 43/84 (51%), Gaps = 5/84 (5%)
Query 99 DQDYRTSLCSTDFPFIEFQSNS-KSGQFFFFSHDGKFLIKTISKAEVIQILRVLPSYIDH 157
++D R +L + +++++ KSG F + D +F++K ++ ++ P Y ++
Sbjct 1554 EEDARIALARSLCKSVQWEARGGKSGSRFCKTLDDRFVLKEMNSRDMTIFEPFAPKYFEY 1613
Query 158 I----LSTPLSLLTRIFGIHRVEL 177
I +LL +IFG+ RV +
Sbjct 1614 IDRCQQQQQPTLLAKIFGVFRVSV 1637
> Hs4753163
Length=3144
Score = 29.3 bits (64), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 33/73 (45%), Gaps = 6/73 (8%)
Query 134 FLIKTISKAEVIQILR---VLPSYIDHILSTPLSLLTR---IFGIHRVELYTTNEAAAAA 187
L KT+ E I + + VL Y+D +L TP +L R I RVE+ ++
Sbjct 1971 MLKKTLQCLEGIHLSQSGAVLTLYVDRLLCTPFRVLARMVDILACRRVEMLLAANLQSSM 2030
Query 188 AAAAADTVQTIQE 200
A + + IQE
Sbjct 2031 AQLPMEELNRIQE 2043
Lambda K H
0.322 0.135 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3682951018
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40