bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0075_orf4
Length=299
Score E
Sequences producing significant alignments: (Bits) Value
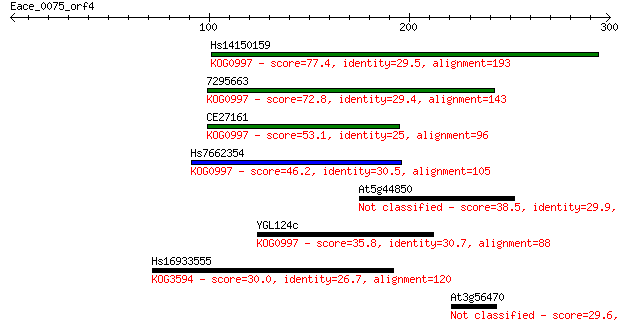
Hs14150159 77.4 4e-14
7295663 72.8 9e-13
CE27161 53.1 7e-07
Hs7662354 46.2 8e-05
At5g44850 38.5 0.016
YGL124c 35.8 0.12
Hs16933555 30.0 6.7
At3g56470 29.6 7.5
> Hs14150159
Length=555
Score = 77.4 bits (189), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 57/193 (29%), Positives = 94/193 (48%), Gaps = 3/193 (1%)
Query 101 GAFEALPLSGTMRHIAMTAVKNSPTPNVLASMLLAEQRVVSLSYAKGYELSPTDVCLLTN 160
GA LPL+ +R +++ + +++ S+LLA ++V+L K L P D+ LL N
Sbjct 291 GAARCLPLAAAVRDTVSASLQQARARSLVFSILLARNQLVALVRRKDQFLHPIDLHLLFN 350
Query 161 FVSSSLALRQLESFTPLCLPSINDRGFLHAYVRYLTPRLAFVCLSSPADSIRFHHLANHC 220
+SSS + R+ E++TP+CLP N GF HA++ YL P L D F +++
Sbjct 351 LISSSSSFREGEAWTPVCLPKFNAAGFFHAHISYLEPDTDLCLLLVSTDREDFFAVSDCR 410
Query 221 NKLFAMLTNTGCLAAIETSLEYAPYALPRCLWGEIALVHCALFVPSLSQYFSSAFGDDYK 280
+ L G A+ +L Y++ + G L H L+ S F+S +
Sbjct 411 RRFQERLRKRGAHLALREALRTPYYSVAQV--GIPDLRHF-LYKSKSSGLFTSPEIEAPY 467
Query 281 TDLSKQQRVYTLY 293
T +Q+R+ LY
Sbjct 468 TSEEEQERLLGLY 480
> 7295663
Length=528
Score = 72.8 bits (177), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 42/145 (28%), Positives = 74/145 (51%), Gaps = 2/145 (1%)
Query 99 ISGAFEALPLSGTMRHIAMTAVKN--SPTPNVLASMLLAEQRVVSLSYAKGYELSPTDVC 156
++ + PL T+R +A+++ S N++ ++L+A ++++L K Y + P D+
Sbjct 257 LTNSIRVFPLPTTIRSQITSAIQSNCSKIKNLVFAVLIANNKLIALVRMKKYSIHPADLR 316
Query 157 LLTNFVSSSLALRQLESFTPLCLPSINDRGFLHAYVRYLTPRLAFVCLSSPADSIRFHHL 216
L+ N V S + + E+++P+CLP + G+LHA+V YL L D F L
Sbjct 317 LIFNLVECSESFKSSENWSPICLPKFDMNGYLHAHVSYLADDCQACLLLLSVDRDAFFTL 376
Query 217 ANHCNKLFAMLTNTGCLAAIETSLE 241
A K+ L + CL AI L+
Sbjct 377 AEAKAKITEKLRKSHCLEAINEELQ 401
> CE27161
Length=479
Score = 53.1 bits (126), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 24/100 (24%), Positives = 52/100 (52%), Gaps = 4/100 (4%)
Query 99 ISGAFEALPLSGTMRHIAMTAVKN----SPTPNVLASMLLAEQRVVSLSYAKGYELSPTD 154
+ + A+P++ + R T + + + L +++A +++ ++ K Y + P D
Sbjct 202 VDSSISAIPMNPSDREFLSTTMASCLGAAKLDGALFGIMIARRQIAAMVRFKKYMIHPRD 261
Query 155 VCLLTNFVSSSLALRQLESFTPLCLPSINDRGFLHAYVRY 194
+ ++ N VS + +++ P+CLP ND GF +AY+ Y
Sbjct 262 LNIVINLVSDNTLQTDSQNWVPICLPRFNDTGFFYAYISY 301
> Hs7662354
Length=547
Score = 46.2 bits (108), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 32/109 (29%), Positives = 53/109 (48%), Gaps = 6/109 (5%)
Query 91 EQQSAALSISGAFEALPLSGTMRHIAMTAVKNSPTPNVLASMLLAEQRVVSLSYAKGY-- 148
EQ AL + GA +PL+ +R ++ P + S+L R+++ + +
Sbjct 237 EQDPGALLL-GAVRCVPLARPLRDALGALLRRCTAPGLALSVLAVGGRLITAAQERNVLA 295
Query 149 --ELSPTDVCLLTNFVSSSLALRQLESFTPLCLPSINDRGFLHAYVRYL 195
L P D+ LL ++V + A E++ P+CLP N GF +AYV L
Sbjct 296 ECRLDPADLQLLLDWVGAP-AFAAGEAWAPVCLPRFNPDGFFYAYVARL 343
> At5g44850
Length=331
Score = 38.5 bits (88), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 40/78 (51%), Gaps = 4/78 (5%)
Query 175 TPLCLPSINDRGFLHAYVRYLTPRLAFVCLSSPADSIRFHHLANHCNKLFAMLTNTGCLA 234
+P CLPS+ ++ + + F+ + +PA +++ HL N C+K + ++ N CL
Sbjct 104 SPACLPSLKKLNLINVVYKDDASLIRFLVIDTPA--VKYIHLTN-CSKQYCLIKNMPCLD 160
Query 235 AIETSLE-YAPYALPRCL 251
S+ YA LPR L
Sbjct 161 RAYISVHTYADDKLPRSL 178
> YGL124c
Length=644
Score = 35.8 bits (81), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 27/98 (27%), Positives = 49/98 (50%), Gaps = 11/98 (11%)
Query 124 PTPNVLASMLLAEQ-RVVSLSYAKGYELSPTDVCLLTNFVS---SSLALRQLESFTPLCL 179
P +L +++A Q ++ + +G+ L TD+ LL +S +L Q E + P+C
Sbjct 353 PRGTLLYGLIIAPQNKLCCVLRPRGHTLHTTDLHLLFCLISHQFQNLDETQ-ELWVPICF 411
Query 180 PSINDRGFLHAYVRYL------TPRLAFVCLSSPADSI 211
P N GFL+ Y+++L + A V +S+ D+
Sbjct 412 PKFNSSGFLYCYIKFLPNDTHSNEKSALVLISAQKDAF 449
> Hs16933555
Length=1955
Score = 30.0 bits (66), Expect = 6.7, Method: Composition-based stats.
Identities = 32/123 (26%), Positives = 53/123 (43%), Gaps = 11/123 (8%)
Query 72 GAATPSTAAEQLHQQQQQQEQQSAALSISGAFEALPLSGTMRH-IAMTAVKNSPTPNVLA 130
G++ P E+ + ++++ +Q L I G + LS S P V
Sbjct 1620 GSSNPLLTTEEANLTEKEEIRQGETLMIEGTEQLKSLSSDSSFCFPRPHFSFSTLPTVSR 1679
Query 131 SM-LLAEQRVVSLSYAKGYELSPTDVCLLTNFVSSSLALRQLESFTPLC-LPSINDRGFL 188
++ L +E V+S ELSP+ C+L SSL+ R+ TP+C LP +R
Sbjct 1680 TVELKSEPNVISSPAECSLELSPSRPCVL----HSSLSRRE----TPICMLPIETERNIF 1731
Query 189 HAY 191
+
Sbjct 1732 ENF 1734
> At3g56470
Length=367
Score = 29.6 bits (65), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 13/22 (59%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 221 NKLFAMLTNTGCLAAIETSLEY 242
N +F LTNTGCLA + SL Y
Sbjct 209 NGVFYCLTNTGCLALFDPSLNY 230
Lambda K H
0.322 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6743279066
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40