bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0068_orf1
Length=292
Score E
Sequences producing significant alignments: (Bits) Value
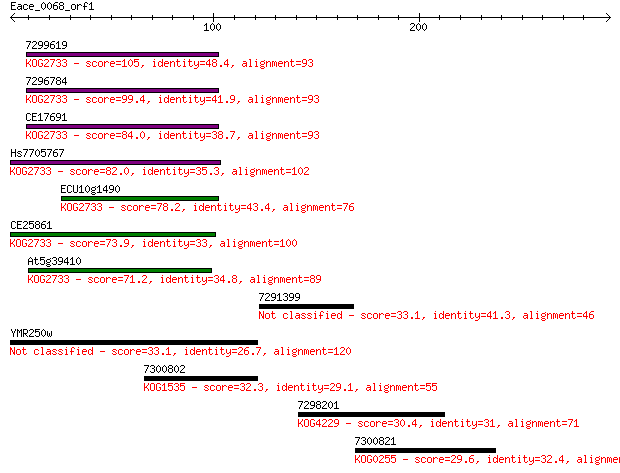
7299619 105 1e-22
7296784 99.4 7e-21
CE17691 84.0 3e-16
Hs7705767 82.0 1e-15
ECU10g1490 78.2 2e-14
CE25861 73.9 4e-13
At5g39410 71.2 2e-12
7291399 33.1 0.64
YMR250w 33.1 0.65
7300802 32.3 1.2
7298201 30.4 4.4
7300821 29.6 7.2
> 7299619
Length=431
Score = 105 bits (262), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 45/93 (48%), Positives = 65/93 (69%), Gaps = 0/93 (0%)
Query 9 VADVDDYKSLLDFTKRCRVLLTTVGPYMLYGEPVARACVETRTHYCDLTGEMPFVALLHA 68
+ADV++ SLL+ KRCR+L+ T GPY +GE V RAC+E TH+ D++GE ++ +
Sbjct 66 IADVNNEASLLEMAKRCRILVNTAGPYRFFGERVVRACIEAGTHHVDVSGEPQYMETMQL 125
Query 69 RHGLAAKERGVKLISFCGFDSIPSDIAVMLIQN 101
R+ AKERGV +IS CGFDSIP+D+ V ++
Sbjct 126 RYHDLAKERGVYVISACGFDSIPADMGVTFVEK 158
> 7296784
Length=430
Score = 99.4 bits (246), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 39/93 (41%), Positives = 66/93 (70%), Gaps = 0/93 (0%)
Query 9 VADVDDYKSLLDFTKRCRVLLTTVGPYMLYGEPVARACVETRTHYCDLTGEMPFVALLHA 68
+ADV+D SLL+ K+CR+++ T GPY +GE V + C+E+ TH+ D++GE ++ +
Sbjct 66 IADVNDQASLLEMAKKCRIVVNTAGPYRFHGENVVKCCIESGTHHVDVSGEPQYMETMQL 125
Query 69 RHGLAAKERGVKLISFCGFDSIPSDIAVMLIQN 101
R+ A+E+GV ++S CGFDSIP+D+ V+ ++
Sbjct 126 RYDQLAREKGVYVVSACGFDSIPADMGVVFVEK 158
> CE17691
Length=426
Score = 84.0 bits (206), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 36/93 (38%), Positives = 59/93 (63%), Gaps = 0/93 (0%)
Query 9 VADVDDYKSLLDFTKRCRVLLTTVGPYMLYGEPVARACVETRTHYCDLTGEMPFVALLHA 68
VAD D +SL + ++ V++ VGPY LYGE V +A VE + D++GE ++ +
Sbjct 66 VADSADERSLNEMARQANVVINAVGPYRLYGEAVVKAAVENGASHVDISGEPAWIEKMQQ 125
Query 69 RHGLAAKERGVKLISFCGFDSIPSDIAVMLIQN 101
++ AKE+GV ++S CG+DSIP+D+ V ++
Sbjct 126 KYSKQAKEQGVYVVSACGWDSIPADLGVNFLKK 158
> Hs7705767
Length=428
Score = 82.0 bits (201), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 36/102 (35%), Positives = 62/102 (60%), Gaps = 0/102 (0%)
Query 1 IEADIHTAVADVDDYKSLLDFTKRCRVLLTTVGPYMLYGEPVARACVETRTHYCDLTGEM 60
+ +++ + D+ + SL + K+ V+L VGPY YGEPV +AC+E D++GE
Sbjct 67 LSSEVGIIICDIANPASLDEMAKQATVVLNCVGPYRFYGEPVIKACIENGASCIDISGEP 126
Query 61 PFVALLHARHGLAAKERGVKLISFCGFDSIPSDIAVMLIQNR 102
F+ L+ ++ A ++GV +I GFDSIP+D+ V+ +N+
Sbjct 127 QFLELMQLKYHEKAADKGVYIIGSSGFDSIPADLGVIYTRNK 168
> ECU10g1490
Length=392
Score = 78.2 bits (191), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 33/76 (43%), Positives = 53/76 (69%), Gaps = 0/76 (0%)
Query 26 RVLLTTVGPYMLYGEPVARACVETRTHYCDLTGEMPFVALLHARHGLAAKERGVKLISFC 85
RVL+ VGPY+ +GE + ++C+ THY D++GE+ F L+ +++ A +GV +I+ C
Sbjct 76 RVLINCVGPYIHHGESIVKSCIRNGTHYMDISGEVYFFELIISKYHDEATRKGVYIINCC 135
Query 86 GFDSIPSDIAVMLIQN 101
GFDSIP DI VM +++
Sbjct 136 GFDSIPFDIGVMCLRD 151
> CE25861
Length=388
Score = 73.9 bits (180), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 33/100 (33%), Positives = 59/100 (59%), Gaps = 0/100 (0%)
Query 1 IEADIHTAVADVDDYKSLLDFTKRCRVLLTTVGPYMLYGEPVARACVETRTHYCDLTGEM 60
++ I V D + +S+ +R ++++ VGP+ L+GE V +A VE + D+ GE
Sbjct 60 LKTRIGLLVCDSTNEESMGKMARRAKLIVNAVGPFRLHGEAVVKAAVENGANQIDVAGEP 119
Query 61 PFVALLHARHGLAAKERGVKLISFCGFDSIPSDIAVMLIQ 100
++ + A++G AK V ++S CG+DSIP+D V L++
Sbjct 120 EWIERMEAKYGQMAKNNNVYIVSACGWDSIPADFGVKLLK 159
> At5g39410
Length=454
Score = 71.2 bits (173), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 31/89 (34%), Positives = 54/89 (60%), Gaps = 0/89 (0%)
Query 10 ADVDDYKSLLDFTKRCRVLLTTVGPYMLYGEPVARACVETRTHYCDLTGEMPFVALLHAR 69
AD D SL + +++L VGP+ ++G+PV AC ++ Y D++GE F+ + A
Sbjct 77 ADTSDPDSLRRLCTQTKLILNCVGPFRIHGDPVVSACADSGCDYLDISGEPEFMERMEAN 136
Query 70 HGLAAKERGVKLISFCGFDSIPSDIAVML 98
+ A+E G ++S CGFDSIP+++ ++
Sbjct 137 YHDRAEETGSLIVSACGFDSIPAELGLLF 165
> 7291399
Length=1024
Score = 33.1 bits (74), Expect = 0.64, Method: Composition-based stats.
Identities = 19/46 (41%), Positives = 26/46 (56%), Gaps = 3/46 (6%)
Query 122 RGGISGATLASALNVIGHKAMNSPYYLTAHAVDGPHNSLAGLPVQP 167
R + G +L SA++ +GH AMN YL+ HA G NS + QP
Sbjct 624 RAEMRGGSLGSAISALGHAAMNG--YLSQHAPLG-QNSSSSAGSQP 666
> YMR250w
Length=585
Score = 33.1 bits (74), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 32/124 (25%), Positives = 53/124 (42%), Gaps = 18/124 (14%)
Query 1 IEADIHTAVADVDDYKSLLDFTKRCRVLLTTVGPYMLYGEPVARACVETRTHYCDLTGEM 60
I+ ++ +AD D+Y L++ T+RC +L + EP+ C TG
Sbjct 106 IDENLTKNLADNDEYPQLIELTQRCISMLAQLWHANPDEEPIG----------CATTGSS 155
Query 61 PFVALLHARHGLAAKERGVKLISFCGFDSIPSDIAVMLIQNRAIEKTKK----PCREVKL 116
+ L GLA K+R + G D+ +I + A+EK + CR V +
Sbjct 156 EAIML----GGLAMKKRWEHRMKNAGKDASKPNIIMSSACQVALEKFTRYFEVECRLVPV 211
Query 117 AVRS 120
+ RS
Sbjct 212 SHRS 215
> 7300802
Length=288
Score = 32.3 bits (72), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 16/56 (28%), Positives = 33/56 (58%), Gaps = 2/56 (3%)
Query 66 LHARHGLAAKERGVKLISFCGFDSIPSDIAVMLIQNRAIEK-TKKPCREVKLAVRS 120
L R GL ++++ L+ F G + +PSD+ +++ Q+ +E+ KK ++ +L V
Sbjct 12 LAKRLGLLSEDQK-SLVEFAGLEGVPSDLKILIAQDPNLEELAKKAEKQPRLEVND 66
> 7298201
Length=2167
Score = 30.4 bits (67), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 31/71 (43%), Gaps = 4/71 (5%)
Query 141 AMNSPYYLTAHAVDGPHNSLAGLPVQPRVCALHRDADFGYSTFYVMARVNENVVRWTNAL 200
A P ++ V G L GLP+QP + AL R F + V + V +A
Sbjct 341 ATEIPEHINVERVAG----LLGLPIQPLIDALTRRTLFAHGETVVSTLSRDQSVDVRDAF 396
Query 201 LDYRYGREFVY 211
+ YGR FV+
Sbjct 397 VKGIYGRMFVH 407
> 7300821
Length=597
Score = 29.6 bits (65), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 32/69 (46%), Gaps = 6/69 (8%)
Query 169 VCALHRDADFGYSTFYVMARVNENVVRWTNALLDYRYGREFVYY-EMLACSFNLLSCVLC 227
VCA D + +V+ RV E + + YGR FVYY ++ CS L ++C
Sbjct 165 VCA----KDLFVTNVFVVGRVTEVAGSFILGQMGDSYGRRFVYYISVVFCSLGRLGSIMC 220
Query 228 MLCTYTALL 236
+YT L
Sbjct 221 T-SSYTWFL 228
Lambda K H
0.325 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6496143184
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40