bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0016_orf1
Length=158
Score E
Sequences producing significant alignments: (Bits) Value
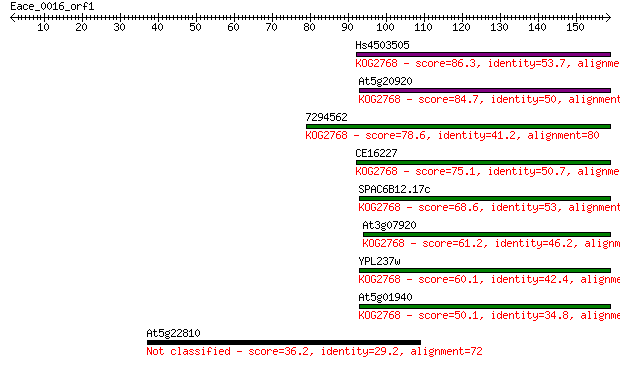
Hs4503505 86.3 2e-17
At5g20920 84.7 6e-17
7294562 78.6 5e-15
CE16227 75.1 6e-14
SPAC6B12.17c 68.6 5e-12
At3g07920 61.2 7e-10
YPL237w 60.1 2e-09
At5g01940 50.1 2e-06
At5g22810 36.2 0.025
> Hs4503505
Length=333
Score = 86.3 bits (212), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 36/69 (52%), Positives = 56/69 (81%), Gaps = 2/69 (2%)
Query 92 SYEEMLSRIQDLIVKNNPDL-AGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPSE 149
+YEE+L+R+ +++ + NPD+ AG KR + +KPPQVVRVG+KK +++NF DIC ++HR +
Sbjct 175 TYEELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKKTSFVNFTDICKLLHRQPK 234
Query 150 HVLQFVLAE 158
H+L F+LAE
Sbjct 235 HLLAFLLAE 243
> At5g20920
Length=268
Score = 84.7 bits (208), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 33/67 (49%), Positives = 53/67 (79%), Gaps = 1/67 (1%)
Query 93 YEEMLSRIQDLIVKNNPDLAGSKRYTI-KPPQVVRVGSKKVAWINFKDICGIMHRPSEHV 151
Y+E+L R+ +++ +NNP+LAG +R T+ +PPQV+R G+KK ++NF D+C MHR +HV
Sbjct 118 YDELLGRVFNILRENNPELAGDRRRTVMRPPQVLREGTKKTVFVNFMDLCKTMHRQPDHV 177
Query 152 LQFVLAE 158
+Q++LAE
Sbjct 178 MQYLLAE 184
> 7294562
Length=312
Score = 78.6 bits (192), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 33/82 (40%), Positives = 54/82 (65%), Gaps = 2/82 (2%)
Query 79 DGSGQLFIRGHVCSYEEMLSRIQDLIVKNNPDLAGSKR--YTIKPPQVVRVGSKKVAWIN 136
D S F +Y+E+L R+ ++I+ NPD+A ++ + ++PPQV+RVG+KK ++ N
Sbjct 141 DNSSTWFGSDRDYTYDELLKRVFEIILDKNPDMAAGRKPKFVMRPPQVLRVGTKKTSFAN 200
Query 137 FKDICGIMHRPSEHVLQFVLAE 158
F DI +HR +H+L F+LAE
Sbjct 201 FMDIAKTLHRLPKHLLDFLLAE 222
> CE16227
Length=250
Score = 75.1 bits (183), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 34/68 (50%), Positives = 47/68 (69%), Gaps = 1/68 (1%)
Query 92 SYEEMLSRIQDLIVKNNPDLAGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPSEH 150
+YEE L+ + ++ NPD AG K+ + IK P+V R GSKK A+ NF +IC +M R +H
Sbjct 88 TYEEALTLVYQVMKDKNPDFAGDKKKFAIKLPEVARAGSKKTAFSNFLEICRLMKRQDKH 147
Query 151 VLQFVLAE 158
VLQF+LAE
Sbjct 148 VLQFLLAE 155
> SPAC6B12.17c
Length=310
Score = 68.6 bits (166), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 35/67 (52%), Positives = 43/67 (64%), Gaps = 2/67 (2%)
Query 93 YEEMLSRIQDLIVKNNPDLAGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPSEHV 151
Y E+L+R L+ NNP+LAG KR YTI PP V R G KK + N DI MHR +HV
Sbjct 159 YPELLNRFFTLLRTNNPELAGEKRKYTIVPPSVHREG-KKTIFANISDISKRMHRSLDHV 217
Query 152 LQFVLAE 158
+QF+ AE
Sbjct 218 IQFLFAE 224
> At3g07920
Length=169
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 30/70 (42%), Positives = 46/70 (65%), Gaps = 5/70 (7%)
Query 94 EEMLSRIQDLIVKNNPDLAGSKRYTI-KPPQVVR----VGSKKVAWINFKDICGIMHRPS 148
+E+L R+ +++ +N+P+L G TI PPQV+R G+KK ++NF D C MHR
Sbjct 17 QEILRRVFNILRENSPELVGIWLLTIIWPPQVLREETAKGTKKTVFVNFMDYCKTMHRNP 76
Query 149 EHVLQFVLAE 158
+HV+ F+LAE
Sbjct 77 DHVMAFLLAE 86
> YPL237w
Length=285
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 28/69 (40%), Positives = 44/69 (63%), Gaps = 4/69 (5%)
Query 93 YEEMLSRIQDLIVKNNPDLAGSK---RYTIKPPQVVRVGSKKVAWINFKDICGIMHRPSE 149
Y E+LSR +++ NNP+LAG + ++ I PP +R G KK + N +DI +HR E
Sbjct 131 YSELLSRFFNILRTNNPELAGDRSGPKFRIPPPVCLRDG-KKTIFSNIQDIAEKLHRSPE 189
Query 150 HVLQFVLAE 158
H++Q++ AE
Sbjct 190 HLIQYLFAE 198
> At5g01940
Length=231
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 44/67 (65%), Gaps = 2/67 (2%)
Query 93 YEEMLSRIQDLIVKNNPDLAGSK-RYTIKPPQVVRVGSKKVAWINFKDICGIMHRPSEHV 151
Y+E+LS + D + + + +++ + R + PPQ++ G+ V +NF D+C MHR +HV
Sbjct 75 YKELLSMVFDRLREEDVEVSTERPRTVMMPPQLLAEGTITVC-LNFADLCRTMHRKPDHV 133
Query 152 LQFVLAE 158
++F+LA+
Sbjct 134 MKFLLAQ 140
> At5g22810
Length=337
Score = 36.2 bits (82), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 40/76 (52%), Gaps = 4/76 (5%)
Query 37 ATSAASSNISSKQQHQQQQQQQHRGLCVSLFAADAAAAEGFIDGSGQLF----IRGHVCS 92
AT + N+ K Q ++ +G + + A A+AA G+ DG+ +L+ + +
Sbjct 59 ATDFTAENLGFKSYPQAYLSKKAKGKNLLIGANFASAASGYYDGTAKLYSAISLPQQLEH 118
Query 93 YEEMLSRIQDLIVKNN 108
Y++ +SRIQ++ NN
Sbjct 119 YKDYISRIQEIATSNN 134
Lambda K H
0.314 0.126 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2100092188
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40