bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Eace_0681_orf4
Length=746
Score E
Sequences producing significant alignments: (Bits) Value
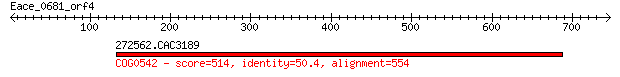
272562.CAC3189 514 5e-144
> 272562.CAC3189
Length=813
Score = 514 bits (1324), Expect = 5e-144, Method: Compositional matrix adjust.
Identities = 279/575 (48%), Positives = 387/575 (67%), Gaps = 31/575 (5%)
Query 133 IGVQHLYAALI---DIPAFQKIMAKSGCDLKIFKSEVVANFKGRQRGV-FPTSFLDMVGD 188
I +H+ ALI + AF I+ G D + + E++ + GR+ TSF
Sbjct 105 INPEHVLLALIRESEGVAFT-ILGNLGVDFERLRKEILDSLSGREENTGIKTSF-----S 158
Query 189 KQGASDVTISQYGVDLSFMAQQGLLPPVIGRDEEIDRIAQILSRKMTKAPMLLGEPGVGK 248
+ T++QYG DL+ MA +G L PVIGRD E RI +IL R+ P L+G+PGVGK
Sbjct 159 SNTSVTPTLNQYGTDLTQMALEGKLDPVIGRDNETQRILEILCRRTKNNPCLIGDPGVGK 218
Query 249 TAVIEGLAQRIVEGAVPESLRNRRIFSLDLLSLSAGSAMRGEYEKRMKEIISYLQAHADE 308
TA+ EGLAQ+IVEG +PE L+++R+ +LD+ SL AGS RGE+E+R+K++++ ++A A
Sbjct 219 TAIAEGLAQKIVEGNIPEILKDKRVITLDVSSLVAGSKYRGEFEERLKKVMTEIKA-AKN 277
Query 309 VILFIDEIHTLIGAGKAAGSMDASQILKVPLARGEIVLVGATTLSEYKLYIEKDAAFCRR 368
VILFIDEIHT++GAG A G++DAS ILK LARGEI +GATT EY+ YIEKDAA RR
Sbjct 278 VILFIDEIHTIVGAGGAEGAIDASNILKPSLARGEIQCIGATTTEEYRRYIEKDAALERR 337
Query 369 FQKIVVEAPSKERTLGILQKVRTKYEDHHNMDIPDEVLAAVVGLSDQYIKRRSFPDKALD 428
FQ + V PSKE + IL+ +R KYE HH + I D+ + A V LS +YI+ R PDKA+D
Sbjct 338 FQPVDVGEPSKEEAVQILEGLRDKYEAHHGVKITDDAIEAAVNLSTRYIQDRFLPDKAID 397
Query 429 LLDESCAMRRVR-------FNNRVAEVGK---------QLED-HRAHRVTLSEEELKELE 471
L+DE+ A R++ N+ E+ K +L+D +A ++ E++L+
Sbjct 398 LIDEAAAKVRIQNLTAPPDLKNQEEELEKTTREKSDAIRLQDFEKAAKLRDKEKKLRIAI 457
Query 472 EEYRTLTEPTDDRPDPDRIELKVDDVARVLSVWTGIPMGKMTEDEMSRILKLADVLGKRV 531
EE++ + T D + + VA V++ WT IP+ K+TE E R+LKL +L KRV
Sbjct 458 EEFKKNWKNTSDN---ENYIVTQKQVAAVVAKWTSIPVEKLTEKESERLLKLESILHKRV 514
Query 532 IGQDEAVQSVANAIRNHRAGLTEEKKPIGAFLFLGSSGVGKTELAKALAEEVFHSEKNLI 591
IGQ+EAV+ V+ A+R R GL + K+PIG+F+FLG +GVGKTEL+KALAE +F E N+I
Sbjct 515 IGQEEAVEYVSRAVRRARVGLKDPKRPIGSFIFLGPTGVGKTELSKALAEAMFGDENNII 574
Query 592 RLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLL 651
R+DM EY E H++SRL+G PPGY+G DEGGQLTE VR+KP+SVVLFDE+E AH +++++L
Sbjct 575 RIDMSEYMEKHTVSRLVGSPPGYVGYDEGGQLTEKVRRKPYSVVLFDEIEKAHPDVFNIL 634
Query 652 LPMLDEGHLTDTKNVRVDFTSTIIIMTSNIGQQYI 686
L +LD+G LTD+K V FT+TIIIMTSN+G I
Sbjct 635 LQILDDGRLTDSKGKTVSFTNTIIIMTSNVGASTI 669
Lambda K H
0.317 0.135 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 352414131850
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40