bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
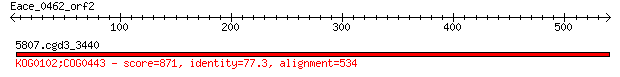
Query= Eace_0462_orf2
Length=540
Score E
Sequences producing significant alignments: (Bits) Value
5807.cgd3_3440 871 0.0
> 5807.cgd3_3440
Length=683
Score = 871 bits (2251), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 413/534 (77%), Positives = 482/534 (90%), Gaps = 0/534 (0%)
Query 6 PKVLENAEGLRTTPSVVAFTKEGERLVGVLAKRQAVTNPENTFFSTKRLIGRQFNEEAIS 65
PKVLEN+EG+RTTPSVVAF+++G+RLVG +AKRQA+TNPENT ++TKRLIGR++ EEAI
Sbjct 73 PKVLENSEGMRTTPSVVAFSEDGQRLVGEVAKRQAITNPENTVYATKRLIGRRYEEEAIK 132
Query 66 KERQILPYKIIRADNGDAWVEAWGKRYSPSQIGAFVLMKMKETAEAYLGRPVGKAVITVP 125
KE+ ILPYKI+RADNGDAWVEA G+RYSPSQIGAF+L KMKETAE YLGR V AVITVP
Sbjct 133 KEQGILPYKIVRADNGDAWVEARGERYSPSQIGAFILEKMKETAETYLGRGVKHAVITVP 192
Query 126 AYFNDSQRQATKDAGRIAGLEVLRIINEPTAAALAYGMEKEDGKTIAVYDLGGGTFDVSL 185
AYFNDSQRQATKDAG IAGL V RIINEPTAAALAYGMEK DGKTIAVYDLGGGTFD+S+
Sbjct 193 AYFNDSQRQATKDAGSIAGLNVTRIINEPTAAALAYGMEKADGKTIAVYDLGGGTFDISI 252
Query 186 LEILGGVFEVKATNGNTSLGGEDFDQKILQYLVQEYQKKEGINLTKDRLALQRLREAAET 245
LEILGGVFEVKATNGNTSLGGEDFDQ+IL YL+QE++K +GI+L++D+LALQRLREA+ET
Sbjct 253 LEILGGVFEVKATNGNTSLGGEDFDQRILNYLIQEFKKTQGIDLSRDKLALQRLREASET 312
Query 246 AKIELSSKLSTEINLPFITADAHGPKHLQISLTRAQLEQLVQPLLQQSVEPCEKCIRDAG 305
AK ELSSK EINLPFITADA GPKHLQI L+RA+ E+LV LL++++ P EKCIRD+G
Sbjct 313 AKKELSSKTQVEINLPFITADARGPKHLQIKLSRAKYEELVDDLLKKTISPSEKCIRDSG 372
Query 306 ISKEDINDIILVGGMTRMPKVADVVKSIFNKEPSKGVNPDEAVAAGAAIQAGVLKGEIKD 365
I KE IND+ILVGGMTRMPKV++ VK IF +EPSKGVNPDEAVA GAAIQAGVLKGEIKD
Sbjct 373 IPKEKINDVILVGGMTRMPKVSETVKKIFGREPSKGVNPDEAVAMGAAIQAGVLKGEIKD 432
Query 366 LLLLDVCPLSLGIETLGGVFTRLINRNTTIPTKKSQIFSTAADNQTQVGIKVYQGEREMA 425
LLLLDV PLSLGIETLGGVFTRLINRNTTIPTKKSQ+FSTAADNQTQVGIKV+QGERE A
Sbjct 433 LLLLDVTPLSLGIETLGGVFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVFQGEREFA 492
Query 426 AANKLLGQFDLVGIPPAPRGVPQIEVIFDVDANGIMNISAIDKTTGKRQQITIQSSGGLN 485
A NKLLGQF+++GIPPAPRGVPQIEV FD+DANGIMN+ AIDK+TGK+ +ITIQSSGGL+
Sbjct 493 ADNKLLGQFEMMGIPPAPRGVPQIEVTFDIDANGIMNVGAIDKSTGKKHEITIQSSGGLS 552
Query 486 EAQIQQMVQDAEKFKEEDKRQKEIIAAKNDAEALIYAVEKQIADLKDKITEEDK 539
A+I++M+++AE+++ D+ +KE+I KNDAEA IY+V+ QI+ L D+I ++K
Sbjct 553 GAEIEKMIREAEEYRANDQAKKELIDLKNDAEAFIYSVQNQISSLADQINTQEK 606
Lambda K H
0.314 0.133 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 238167410591
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40