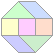
 Eten_3138 (170 letters)
Eten_3138 (170 letters)
9 hits
 170 letters
170 letters
cel 



 169.0 - CE10362 (227) KOG1689
169.0 - CE10362 (227) KOG1689
dme 



 165.0 - 7294949 (237) KOG1689
165.0 - 7294949 (237) KOG1689
hsa 



 155.0 - Hs5901926 (227) KOG1689
155.0 - Hs5901926 (227) KOG1689
ath 



 155.0 - At4g29820 (185) KOG1689
155.0 - At4g29820 (185) KOG1689
ath 



 146.0 - At4g25550 (210) KOG1689
146.0 - At4g25550 (210) KOG1689
cel 



 73.0 - CE15213 (1170) KOG1023
73.0 - CE15213 (1170) KOG1023
ath 



 62.0 - At5g23230 (198)
62.0 - At5g23230 (198)
ath 



 62.0 - At5g34960_1 (591) KOG0851
62.0 - At5g34960_1 (591) KOG0851
hsa 



 62.0 - Hs22053744 (679)
62.0 - Hs22053744 (679)
BLASTP 2.2.18 Mar-02-2008
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query: Eten_3138 (170 letters)
Database: /usr/local/genome/databases/kog/kyva
47500486 sequences; 112920 total letters
>gnl|BL_ORD_ID|50118 CE10362
Length = 227
Score: 169.0 Expect: 2.39775e-12
Identities 32 (40%) Positives: 45 (56%) Gaps: 2/80 (2%)
Query: 19 LGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRCSFRIPVGFCLTVVPIFEL--SPQRVGLAIGGLSHLIARFSI 96
+G WWR FD PY+P HVT+PKE ++ VQLP + +F +P F L P+FEL + G I L ++RF+
Sbjct: 142 IGNWWRPNFDPPRYPYIPAHVTKPKEHTKLLLVQLPSKSTFCVPKNFKLVAAPLFELYDNAAAYGPLISSLPTTLSRFNF 221
>gnl|BL_ORD_ID|9024 7294949
Length = 237
Score: 165.0 Expect: 6.97639e-12
Identities 33 (37%) Positives: 47 (54%) Gaps: 2/87 (2%)
Query: 15 VGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRCSFRIPVGFCLTVVPIFEL--SPQRVGLAIGGLSHLIARFSIRLM 99
V + +G WWR F+ PY+PPH+T+PKE R+ VQL + F +P + L P+FEL + Q G I L + RF+ M
Sbjct: 151 VEDTIGNWWRPNFEPPQYPYIPPHITKPKEHKRLFLVQLHEKALFAVPKNYKLVAAPLFELYDNSQGYGPIISSLPQALCRFNFIYM 237
>gnl|BL_ORD_ID|97475 Hs5901926
Length = 227
Score: 155.0 Expect: 1.1228e-10
Identities 30 (34%) Positives: 46 (52%) Gaps: 2/87 (2%)
Query: 12 DLEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRCSFRIPVGFCLTVVPIFELSPQRVGLA--IGGLSHLIARFSI 96
D + + +G WWR F+ PY+P H+T+PKE ++ VQL + F +P + L P+FEL G I L L++RF+
Sbjct: 138 DWVIDDCIGNWWRPNFEPPQYPYIPAHITKPKEHKKLFLVQLQEKALFAVPKNYKLVAAPLFELYDNAPGYGPIISSLPQLLSRFNF 224
>gnl|BL_ORD_ID|35803 At4g29820
Length = 185
Score: 155.0 Expect: 1.13221e-10
Identities 31 (32%) Positives: 55 (57%) Gaps: 2/96 (2%)
Query: 6 LSTDGPDLEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRCSFRIPVGFCLTVVPIFEL--SPQRVGLAIGGLSHLIARFSIRLM 99
L++ ++ VGE +G WWR F+ + P+LPP++ PKE ++ V+LP F +P F L VP+ +L + + G + + L+++FS +M
Sbjct: 88 LASLCINIAVGECIGMWWRPNFETLMYPFLPPNIKHPKECTKLFLVRLPVHQQFVVPKNFKLLAVPLCQLHENEKTYGPIMSQIPKLLSKFSFNMM 183
>gnl|BL_ORD_ID|35384 At4g25550
Length = 210
Score: 146.0 Expect: 1.25181e-09
Identities 29 (40%) Positives: 39 (54%) Gaps: 6/71 (8%)
Query: 11 PDLEVGEFLGEWWRGEFDDDLVPYLPPHVTRPK------ERVRVHQVQLPHRCSFRIPVGFCLTVVPIFEL 75
PD VGE + WWR F+ + PY PPH+T+PK E R++ V L + F +P L VP+FEL
Sbjct: 123 PDWTVGECVATWWRPNFETMMYPYCPPHITKPKVVKKHNECKRLYIVHLSEKEYFAVPKNLKLLAVPLFEL 193
>gnl|BL_ORD_ID|51937 CE15213
Length = 1170
Score: 73.0 Expect: 0.365066
Identities 17 (36%) Positives: 27 (57%) Gaps: 7/47 (14%)
Query: 125 TSFDIHELEEQPDV------AGGHSEAKPALENDGELEMLPPELQHL 165
++FD+ +E+PDV GGH+ +P+L+ GE + P L HL
Sbjct: 792 SAFDLENRKEKPDVIIYQVKKGGHNPMRPSLDT-GETVEINPALLHL 837
>gnl|BL_ORD_ID|38737 At5g23230
Length = 198
Score: 62.0 Expect: 5.97437
Identities 13 (37%) Positives: 18 (51%) Gaps: 6/35 (17%)
Query: 18 FLGEWWRGEF------DDDLVPYLPPHVTRPKERV 46
LGEWW G+ D +++P + VT P E V
Sbjct: 72 MLGEWWNGDLILDGTTDSEIIPEINRQVTGPDEIV 106
>gnl|BL_ORD_ID|1182 At5g34960_1
Length = 591
Score: 62.0 Expect: 6.49424
Identities 19 (30%) Positives: 28 (45%) Gaps: 1/62 (1%)
Query: 94 FSIRLMVPTLDKYEADTSLEHKGEAGLDDLGTSFDIHELEEQPDVAGGHSEAKPALENDGEL 155
FS + +P L SL+ AG+ L SFD ++ PD H ++ L NDG +
Sbjct: 162 FSFFVPIPFLSILHDLWSLKCYETAGVKSLSNSFDASQVHVNPDFPEAHHFSQ-TLPNDGAI 222
>gnl|BL_ORD_ID|89655 Hs22053744
Length = 679
Score: 62.0 Expect: 7.48395
Identities 11 (32%) Positives: 18 (52%) Gaps: 0/34 (0%)
Query: 113 EHKGEAGLDDLGTSFDIHELEEQPDVAGGHSEAK 146
+H+ + G+ G S+ +H QP G HSE +
Sbjct: 54 DHRAQQGVQRQGVSYSVHAYTGQPSPRGLHSENR 87
Database: /usr/local/genome/databases/kog/kyva
Number of letters in database: 47500486
Number of sequences in database: 112920
Matrix: BLOSUM62
Gap penalties: Existence: 11 Extension: 1
Number of sequences: 112920
Length of database: 47500486
Effecttive HSP length: 0
Effecttive length of query: 170
Effective search space: 0.0
Effective search space used: 0.0
T: 11
A: 40
X1: 16 (7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (20.4 bits)
S2: 73 (32.7 bits)
 Eten_3138 (170 letters)
Eten_3138 (170 letters) Eten_3138 (170 letters)
Eten_3138 (170 letters) 170 letters
170 letters



 169.0 - CE10362 (227) KOG1689
169.0 - CE10362 (227) KOG1689



 165.0 - 7294949 (237) KOG1689
165.0 - 7294949 (237) KOG1689



 155.0 - Hs5901926 (227) KOG1689
155.0 - Hs5901926 (227) KOG1689



 155.0 - At4g29820 (185) KOG1689
155.0 - At4g29820 (185) KOG1689



 146.0 - At4g25550 (210) KOG1689
146.0 - At4g25550 (210) KOG1689



 73.0 - CE15213 (1170) KOG1023
73.0 - CE15213 (1170) KOG1023



 62.0 - At5g23230 (198)
62.0 - At5g23230 (198)



 62.0 - At5g34960_1 (591) KOG0851
62.0 - At5g34960_1 (591) KOG0851



 62.0 - Hs22053744 (679)
62.0 - Hs22053744 (679)